ABSTRACT
The capture of live bait for sport fishing is an important activity for fishing communities. The main species used for this purpose are members of the genus Gymnotus, which comprises numerous species of cryptic nature that are difficult to identify based on external morphology. The aims of this work were to identify through partial sequences of the COI gene Gymnotus species fished in the Jacaré-Guaçu River, SP, and to develop a molecular diagnostic approach using PCR-RFLP to identify these species. Partial COI sequences were compared to those of other species deposited in GenBank. The sequences were assessed in the NEBCutter program to determine restriction sites in the sequence and the enzymes to be tested. Phenetic analysis performed by Neighbor-Joining method showed that the specimens sampled belong to two species preliminary identified here as G. cf. sylvius and G. cf. cuia, with G. cf. sylvius accounting for 95.2% of the individuals sampled. The enzymes NlaIII and SacI generated fragments that allowed distinguishing the Gymnotus species using PCR-RFLP. This analysis can be used to accurately identify these species, which is fundamental for monitoring Gymnotus fishing and assessing the conservation of this genetic resource.
Keywords:
Molecular identification; PCR-RFLP; Tuvira
RESUMO
A captura de iscas-vivas para a pesca esportiva constitui uma atividade importante em comunidades de pescadores. As principais espécies utilizadas para este propósito pertencem ao gênero Gymnotus, o qual compreende inúmeras espécies de natureza críptica que dificulta a identificação baseada na morfologia externa. Os objetivos deste trabalho foram identificar através de sequências parciais do gene COI, espécies de Gymnotus capturadas no Rio Jacaré-Guaçu, Ibitinga, SP, e desenvolver um diagnóstico molecular por meio de PCR-RFLP. Sequências parciais de COI foram comparadas com outras espécies depositadas no GenBank. As sequências foram analisadas no Programa NebCutter para determinar os sítios de restrição e definir as enzimas a serem testadas. A análise fenética pelo método de Neighbor-Joining mostrou que os espécimes pertencem a duas espécies identificadas preliminarmente aqui como G. cf. sylvius e G. cf. cuia, sendo que G. cf. sylvius representou 95,2% dos indivíduos amostrados. As enzimas NlaIII e SacI geraram fragmentos que permitiram discriminar as espécies por meio de PCR-RFLP. Esta análise pode ser usada na identificação precisa destas espécies, fundamental na proposição de monitoramento da pesca de Gymnotus na região e para medidas adequadas de conservação.
Palavras-chave:
Identificação molecular; PCR-RFLP; Tuvira
Introduction
Species of Gymnotus Linnaeus, 1758 are an important fishing resource for fishermen communities and the principal source of commercial live bait for sport fishing (Sousa et al., 2017Sousa TP, Marques DKS, Vitorino CA, Faria KC, Braga GSF, Ferreira DC, Venere PC. Cytogenetic and molecular data support the occurrence of three Gymnotus Species (Gymnotiformes: Gymnotidae) used as live bait in Corumbá, Brazil: implications for conservation and management of professional fishing. Zebrafish. 2017; 14(2):177-86. https://doi.org/10.1089/zeb.2016.1356
https://doi.org/10.1089/zeb.2016.1356...
). The genus Gymnotus is a member of the family Gymnotidae (order Gymnotiformes). This genus consists of many species, represented by 43 valid species in six clades (Tagliacollo et al., 2016Tagliacollo VA, Bernt MJ, Craig JM, Oliveira C, Albert JS. Model-based total evidence phylogeny of Neotropical electric knifefishes (Teleostei, Gymnotiformes). Mol Phylogenet Evol. 2016; 95:20-33. https://doi.org/10.1016/j.ympev.2015.11.007
https://doi.org/10.1016/j.ympev.2015.11....
), with wide distribution in the continental waters of South and Central America, except in Chile and Belize (Albert, 2001Albert JS. Species diversity and phylogenetic systematics of American knifefishes (Gymnotiformes, Teleostei). Misc Publ Mus Zool Univ Mich. 2001; 190:1-127.).
The identification of species in this genus based on morphological characters is sometimes challenging due to the morphological conservatism among species and high phenotypic plasticity within species. Many efforts have been made based on studies of morphology, cytogenetics, molecular, and electrical signals to elucidate the taxonomy of electric fishes, including Gymnotus (Alves-Gomes et al., 1995Alves-Gomes JA, Ortí G, Haygood M, Heiligenberg W, Meyer A. Phylogenetic analysis of the South American electric fishes (Order Gymnotiformes) and the evolution of their electrogenic system: A synthesis based on morphology, electrophysiology, and mitochondrial sequence data. Mol Biol Evol. 1995; 12(2):298-318. https://doi.org/10.1093/oxfordjournals.molbev.a040204
https://doi.org/10.1093/oxfordjournals.m...
; Albert, Crampton, 2003Albert JS, Crampton WGR. Seven new species of the Neotropical electric fish Gymnotus (Teleostei, Gymnotiformes) with a redescription of G. carapo (Linnaeus). Zootaxa. 2003; 287(1):1-54. http://dx.doi.org/10.11646/zootaxa.287.1.1
http://dx.doi.org/10.11646/zootaxa.287.1...
; Fernandes et al., 2005Fernandes FMC, Albert JS, Daniel-Silva MFZ, Lopes CE, Crampton WGR, Almeida-Toledo LF. A new Gymnotus (Teleostei: Gymnotiformes: Gymnotidae) from the Pantanal Matogrossense of Brazil and adjacent drainages: Continued documentation of a cryptic fauna. Zootaxa . 2005; 933(1):1-14. http://dx.doi.org/10.11646/zootaxa.933.1.1
http://dx.doi.org/10.11646/zootaxa.933.1...
; Margarido et al., 2007Margarido VP, Bellafronte E, Moreira-Filho O. Cytogenetic analysis of three sympatric Gymnotus (Gymnotiformes, Gymnotidae) species verifies invasive species in the Upper Paraná River basin, Brazil. J Fish Biol. 2007; 70(sb):155-64. https://doi.org/10.1111/j.1095-8649.2007.01365.x
https://doi.org/10.1111/j.1095-8649.2007...
; Maxime, Albert, 2009Maxime EL, Albert JS. A new species of Gymnotus (Gymnotiformes: Gymnotidae) from the Fitzcarrald Arch of southeastern Peru. Neotrop Ichthyol. 2009; 7(4):579-85. http://dx.doi.org/10.1590/S1679-62252009000400004
http://dx.doi.org/10.1590/S1679-62252009...
; Scacchetti et al., 2011Scacchetti PC, Pansonato-Alves JC, Utsunomia R, Oliveira C, Foresti F. Karyotypic diversity in four species of the genus Gymnotus Linnaeus, 1758 (Teleostei, Gymnotiformes, Gymnotidae): Physical mapping of ribosomal genes and telomeric sequences. Comp Cytogenet. 2011; 5(3):223-35. https://doi.org/10.3897/compcytogen.v5i3.1375
https://doi.org/10.3897/compcytogen.v5i3...
; Milhomem et al., 2012Milhomem SSR, Crampton WGR, Pieczarka JC, Shetka GH, Silva DS, Nagamachi CY. Gymnotus capanema, a new species of electric knife fish (Gymnotiformes, Gymnotidae) from eastern Amazonia, with comments on an unusual karyotype. J Fish Biol . 2012; 80(4):802-15. https://doi.org/10.1111/j.1095-8649.2012.03219.x
https://doi.org/10.1111/j.1095-8649.2012...
; Tagliacollo et al., 2016Tagliacollo VA, Bernt MJ, Craig JM, Oliveira C, Albert JS. Model-based total evidence phylogeny of Neotropical electric knifefishes (Teleostei, Gymnotiformes). Mol Phylogenet Evol. 2016; 95:20-33. https://doi.org/10.1016/j.ympev.2015.11.007
https://doi.org/10.1016/j.ympev.2015.11....
; Sousa et al., 2017Sousa TP, Marques DKS, Vitorino CA, Faria KC, Braga GSF, Ferreira DC, Venere PC. Cytogenetic and molecular data support the occurrence of three Gymnotus Species (Gymnotiformes: Gymnotidae) used as live bait in Corumbá, Brazil: implications for conservation and management of professional fishing. Zebrafish. 2017; 14(2):177-86. https://doi.org/10.1089/zeb.2016.1356
https://doi.org/10.1089/zeb.2016.1356...
; Craig et al., 2017Craig JM, Crampton WGR, Albert JS. Revision of the polytypic electric fish Gymnotus carapo (Gymnotiformes, Teleostei), with descriptions of seven species. Zootaxa . 2017; 4318(3):401-38. http://dx.doi.org/10.11646/zootaxa.4318.3.1
http://dx.doi.org/10.11646/zootaxa.4318....
, 2018Craig JM, Malabarba LR, Crampton WGR, Albert JS. Revision of banded knifefishes of the Gymnotus carapo and G. tigre clades (Gymnotidae Gymnotiformes) from the Southern Neotropics. Zootaxa 2018; 4379(1):47-73. http://dx.doi.org/10.11646/zootaxa.4379.1.3
http://dx.doi.org/10.11646/zootaxa.4379....
).
The existence of cryptic species in combination with poor-quality data from landing records for biomass estimation and catch quotas, may imperil the long-term survival of given fish genetic resources (Hilsdorf, Hallerman, 2017Hilsdorf AWS, Hallerman EM. Genetic resources of Neotropical fishes. Cham, Switzerland: Springer International Publishing AG; 2017.). In addition, worldwide concerns on the indiscriminate use of live baitfish, the ecological risks of introducing alien species, and the depletion of local species, are on the agenda of fish diversity conservation in many countries (Drake, Mandrak, 2014Drake DAR, Mandrak NE. Ecological risk of live bait fisheries: A new angle on selective fishing. Fisheries 2014; 39(5):201-11. https://doi.org/10.1080/03632415.2014.903835
https://doi.org/10.1080/03632415.2014.90...
; Sousa et al., 2017Sousa TP, Marques DKS, Vitorino CA, Faria KC, Braga GSF, Ferreira DC, Venere PC. Cytogenetic and molecular data support the occurrence of three Gymnotus Species (Gymnotiformes: Gymnotidae) used as live bait in Corumbá, Brazil: implications for conservation and management of professional fishing. Zebrafish. 2017; 14(2):177-86. https://doi.org/10.1089/zeb.2016.1356
https://doi.org/10.1089/zeb.2016.1356...
). Therefore, the correct identification of species is one of the first steps towards successfully establishing conservation and management measures to protect local baitfish species. Hebert et al. (2003Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc B. 2003; 270(1512):313-21. https://doi.org/10.1098/rspb.2002.2218
https://doi.org/10.1098/rspb.2002.2218...
) proposed the use of a fragment of approximately 650bp of the mitochondrial cytochrome c oxidase subunit 1 (COI or COX1) as a global identification system for most animal species presently known as the DNA barcoding.
Supported by this background, we use partial sequences of COI to identify Gymnotus spp. captured as live baitfish and develop a rapid and low-cost molecular methodology to identify unequivocally the species being caught to help ensure the sustainability of Gymnotus fisheries and preserve the biodiversity of species.
Material and Methods
Collection and Sampling Site. Taxon sampling was carried out between October 2015 and March 2016 along the Jacaré-Guaçu River, a tributary of the Tietê River, southeast region of Brazil (Fig. 1). Eight individuals from four fishing sites were collected in two season periods, in a total of 64 individuals. All specimens were photographed, labeled and a small piece of muscle or fin clips were taken from each fresh fish and stored in 95% ethanol at -20°C. Voucher specimens were deposited in the Ichthyology Collection of the Museu de Zoologia da Universidade de São Paulo (MZUSP), SP, Brazil, receiving the register numbers MZUSP 124930 and MZUSP 124931.
Map of the study area. Characterization of the catchment sites of Gymnotus spp. in the Jacaré-Guaçu River, southeastern Brazil. Orange dots (A1-A4) = collection sites; green dot = embarkation and disembarkation; blue rectangle = Ibitinga dam; lines on the diagonal = boundary of the State of São Paulo. 1: 100,000 (UTM projection), WGS Datum 1984. Elaborated by Luís Campanha, modified by Lilian P. Faria Pereira.
The procedures carried out in this study were in accordance with the ethical standards of the Colégio Brasileiro de Experimentação Animal (COBEA) and were approved by the Comitê de Ética em Experimentação Animal do Instituto de Pesca (CEEAIP) protocol number 01/2016.
DNA extraction, PCR and Sequencing. Total genomic DNA was extracted following the protocol of saline extraction method (Aljanabi, Martinez, 1997Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997; 25(22):4692-93. https://doi.org/10.1093/nar/25.22.4692
https://doi.org/10.1093/nar/25.22.4692...
). Partial sequences of the cytochrome c oxidase 1 (COI or Cox1) gene have been amplified by the polymerase chain reaction (PCR) using the universal primers FishF1 (5 ‘ - TCA ACC AAC CAC AAA GAC ATT GGC AC-3 ‘) and FishR1 (5 ‘ - TAG ACT TCT GGG TGG CCA AAG AAT CA-3’) (Ward et al., 2005Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philos Trans R Soc London B. 2005; 360(1462):1847-57. https://doi.org/10.1098/rstb.2005.1716
https://doi.org/10.1098/rstb.2005.1716...
). PCR was conducted for final volume of 50 µL containing 1x buffer solution, 0.2 mM of each DNTP, 25 mM MgCl2, 0.2 µM of each primer (sense and anti-sense), 1.0 U of Platinum Taq DNA Polymerase High Fidelity (InvitrogenTM, Carlsbad, CA, USA) and 0.5 - 2 ng of DNA. The amplification reaction was conducted in a PTC-200 thermal cycler (MJ Research, Waltham, MA, USA) with an initial cycle of denaturation at 94°C for 2 min, followed by 35 cycles (94°C for 40 S, 58°C for 1 min, 72°C for 1 min), and the final extension at 72°C for 7 min.
PCR products were purified with CleanSweep™ PCR Purification Reagent (Applied Biosystems™), according to the protocol provided by the manufacturer. For the sequencing reactions the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems ™) was used. Sequencing was performed on the ABI 3730 DNA Analyzer (Applied Biosystems™).
Sequence Analysis. The COI partial sequences of all samples were edited in the Chromas V.2.6 program. The Codon Code Aligner V 1.5.2 program (Codon Code Corporation, Dedham, Massachusetts, United States) was used for quality evaluation and obtaining the consensus sequence (contig) from the forward and reverse sequences.
The sequences were compared to the sequences of other species of Gymnotus deposited in GenBank (S1 - Available only as online supplementary file accessed with the online version of the article at http://www.scielo.br/ni) for the identification of similarity using the MegaBLAST program (Zhang et al., 2000Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000; 7(1-2):203-14. https://doi.org/10.1089/10665270050081478; Morgulis et al., 2008Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics 2008; 24(16):1757-64. https://doi.org/10.1093/bioinformatics/btn322
https://doi.org/10.1093/bioinformatics/b...
). The number of haplotypes of the partial sequences of the COI gene was determined in the DnaSP program V.5.10 (Rozas et al., 2003Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003; 19(18):2496-97. https://doi.org/10.1093/bioinformatics/btg359
https://doi.org/10.1093/bioinformatics/b...
). Sequences from each haplotype were deposited in GenBank (http://www.ncbi.nlm.nih.gov) (accession numbers: MN167125, MN167126, MN167127, MN167128, MN167129, MN167130, MN167131).
The sequences were aligned with the ClustalW command (Thompson et al., 1994Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res . 1994; 22(22):4673-80. https://doi.org/10.1093/nar/22.22.4673
https://doi.org/10.1093/nar/22.22.4673...
) implemented in the MEGA 6 program (Tamura et al., 2014Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol . 2014; 30(12):2725-29. https://doi.org/10.1093/molbev/mst197
https://doi.org/10.1093/molbev/mst197...
). Neighbor-Joining (NJ) based on Kimura two Parameter (K2P) genetic distance were used to analyze the phenetic relationship between the samples, as recommended by Barcode of Life. The bootstrap re-sampling analysis was performed using 1,000 replicates (Nei, Kumar, 2000Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000.) and the genetic divergence was calculated by the K2P distances (Kimura, 1980Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16(2):111-20.).
PCR-RFLP. Gymnotus partial COI sequences amplified herein, and those retrieved from GenBank were assessed using the NEBCutter program (Vincze et al., 2003Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res . 2003; 31(13):3688-91. https://doi.org/10.1093/nar/gkg526
https://doi.org/10.1093/nar/gkg526...
). The diagnostic enzymes were used to distinguish among the Gymnotu sampled individuals.
Digestion was carried out at 10 µL final volume, including 1.0 µL of endonuclease (10 U.mL-1) (BioLabs), 1 µL of 10x NEBuffer 4 (50mM Potassium Acetate, 20mM Tris-acetate, 10mM Magnesium Acetate, 1mM dithiothreitol (DTT), pH 7.9, 25ºC; Biolabs) and 5 µL of the PCR product. Solutions were incubated for 1-2 h at 37°C in the thermal block. The resulting sizes of restriction fragments were determined in 3.0% agarose gel electrophoresis by comparison with a standard DNA molecular weight marker. Images were digitalized for further analysis of the banding patterns.
Results
Molecular Identification. Out of 64 sequences examined only two showed poor quality and could not be used. After editing, the sequences were 585 bp long. Of the 62 successfully sequenced samples, 59 showed similarity of 99 to 100% with the species G. sylvius Albert & Fernandes-Matioli, 1999 and the other three sequences presented a best identity with G. inaequilabiatus (Valenciennes, 1839) when compared with other GenBank sequences in the MegaBLAST program. However, a species level match was not made, presumably due to misidentifications of the species names in GenBank.
The bootstrap values indicate branch statistical supports. Analysis showed that specimens from the fishing in the Jacaré-Guaçu River grouped in two different clades: one with sequences of G. sylvius (clade A) and another one with sequences of G. inaequilabiatus (clade C) (Fig. 2). The first one comprising 95.2% of the individuals sampled and the second one 4.8%. Specimens grouped in clade A yielded 5 haplotypes (Photos S2https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/82b2c525a42a4976d45e89349fdac33f535f73be.jpg, S7https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/e491b1d9a116ee69a6c99b9556dc2e3e41cab72e.jpg and S8https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/907ad47ddc76a6bb4a0dfc9059f4e877da2cb463.jpg) with 3 polymorphic sites (n = 3).S4https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/af7872c9797f8dd8edad536954e5f79100a309bd.jpg, S6https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/0a558cdf00a346ca6c91717efec172f017a19a8c.jpg) with 4 polymorphic sites (n = 59) and in clade C, 2 haplotypes (S3https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/e79d98de62204f0a24916bd550ac9e4bdf318d57.jpg, S5https://minio.scielo.br/documentstore/1982-0224/NsgQqzBWJyxRSxNfjjvTqgz/33ca0b4cfc6e9b9ceeaa9081f010217fbf2cd27a.jpg and
Neighbor-joining dendrogram of the COI partial sequences of Gymnotus spp. (1T13, 2T29, 1T14, 1T36, 2T26, 2T13, 2T32 and 2T24) collected in the Jacaré-Guaçu River, State of São Paulo, and obtained from GenBank, constructed from the distances of Kimura (K2P). The bootstrap values are indicated on the branches.
Considering a recent revision of Craig et al. (2017Craig JM, Crampton WGR, Albert JS. Revision of the polytypic electric fish Gymnotus carapo (Gymnotiformes, Teleostei), with descriptions of seven species. Zootaxa . 2017; 4318(3):401-38. http://dx.doi.org/10.11646/zootaxa.4318.3.1
http://dx.doi.org/10.11646/zootaxa.4318....
) with the description of seven subspecies of G. carapo Linnaeus, 1758, and a revision of Craig et al. (2018Craig JM, Malabarba LR, Crampton WGR, Albert JS. Revision of banded knifefishes of the Gymnotus carapo and G. tigre clades (Gymnotidae Gymnotiformes) from the Southern Neotropics. Zootaxa 2018; 4379(1):47-73. http://dx.doi.org/10.11646/zootaxa.4379.1.3
http://dx.doi.org/10.11646/zootaxa.4379....
) of G. carapo and G. tigreAlbert & Crampton, 2003Albert JS, Crampton WGR. Seven new species of the Neotropical electric fish Gymnotus (Teleostei, Gymnotiformes) with a redescription of G. carapo (Linnaeus). Zootaxa. 2003; 287(1):1-54. http://dx.doi.org/10.11646/zootaxa.287.1.1
http://dx.doi.org/10.11646/zootaxa.287.1...
clade we preliminary identified here two species: G. cf. sylvius and G. cf. cuia Craig, Malabarba, Crampton & Albert, 2018. Species of G. cf. sylvius were collected in the four fishing sites and G. cf. cuia in A2, A3 and A4.
The intraspecific genetic divergence for G. cf. sylvius ranged from 0.0 to 0.6% and for G. cf. cuia from 0.0 to 0.9%. The divergence between G. cf. sylvius and G. cf. cuia ranged from 4.7 to 5.6% (Tab. 1).
PCR-RFLP. Based on the in silico analysis, we determined the enzymes that can be used for the diagnosis of the Gymnotus species present in the Jacaré-Guaçu River. The enzymes that allowed differentiation between the species were NlaIII (5 ‘... CATG’ ... 3’) and SacI (5’ ... GAGCT’C ... 3 ‘). These two enzymes generated fragments that allowed also to differentiate the species G. pantanal Fernandes, Albert, Daniel-Silva, Lopes, Crampton & Almeida-Toledo, 2005 and G. pantherinus (Steindachner, 1908), in places where these two species occur in sympatry with G. cf. sylvius and G. cf. cuia (Tabs. 2-3).
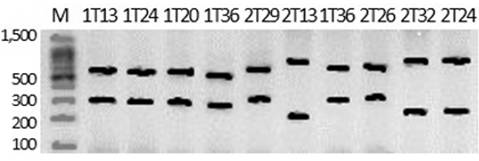
COI cleavage patterns using NlaIII and SacI endonucleases are depicted in the Figs. 3-4.
Agarose gel electrophoresis (3.0%) of NlaIII restriction fragments obtained from COI partial PCR amplification. Gymnotus cf. sylvius (1T13, 1T24, 1T20, 1T36, 2T29) and G. cf. cuia (2T13, 2T32 and 2T24). M- 100 bp molecular weight marker (Sinapse Inc). Numbers at fragments indicate estimated molecular weight.
Agarose gel electrophoresis (3.0%) of NlaIII restriction fragments obtained from COI partial PCR amplification. Gymnotus cf. sylvius (1T13, 1T24, 1T20, 1T36, 2T26, 2T29) and G. cf. cuia (2T13, 2T32 and 2T24). M- 100 bp molecular weight marker (Sinapse Inc). Numbers at fragments indicate estimated molecular weight.
Size of fragments generated by NlaIII for G. cf. sylvius, G. cf. cuia, G. pantherinus and G. pantanal.
Size of fragments generated by SacI for G. cf. sylvius, G. cf. cuia, G. pantherinus and G. pantanal.
Discussion
Successful conservation plans need precise species identification (FAO, 2013Food and Agriculture Organization (FAO). Fish identification tools for biodiversity and fisheries assessments: Review and guidance for decision-makers. Rome: Food and Agriculture Organization of the United Nations [Internet]. Rome; 2013. Available from: http://www.fao.org/docrep/019/i3354e/i3354e.pdf
http://www.fao.org/docrep/019/i3354e/i33...
). Thus, genetic markers have been applied widely for traceability of fish and fish product as well as for the species identification and populations to enforce fisheries regulation for the conservation of fisheries resources (Ogden, 2008Ogden R. Fisheries forensics: The use of DNA tools for improving compliance, traceability and enforcement in the fishing industry. Fish Fish. 2008; 9(4):462-72. https://doi.org/10.1111/j.1467-2979.2008.00305.x
https://doi.org/10.1111/j.1467-2979.2008...
). Partial sequences of COI obtained in this study demonstrated that the Gymnotus species caught as live bait by artisanal fisher community in the Jacaré-Guaçu River belong to two distinct species - G. cf. sylvius and G. cf. cuia.
Species of Gymnotus known to occur in the Upper Paraná basin include: G. carapo australis Craig, Crampton & Albert, 2017, G. cuia, G. inaequilabiatus, G. pantanal, G. pantherinus, G. paraguensis Albert & Crampton, 2003 and G. sylvius (Craig et al., 2017Craig JM, Malabarba LR, Crampton WGR, Albert JS. Revision of banded knifefishes of the Gymnotus carapo and G. tigre clades (Gymnotidae Gymnotiformes) from the Southern Neotropics. Zootaxa 2018; 4379(1):47-73. http://dx.doi.org/10.11646/zootaxa.4379.1.3
http://dx.doi.org/10.11646/zootaxa.4379....
; Craig et al., 2018Drake DAR, Mandrak NE. Ecological risk of live bait fisheries: A new angle on selective fishing. Fisheries 2014; 39(5):201-11. https://doi.org/10.1080/03632415.2014.903835
https://doi.org/10.1080/03632415.2014.90...
). Of these species, G. carapo australis, G. cuia, and G. pantanal are ecologically abundant and G. inaequilabiatus, G. pantherinus, G. paraguensis and G. sylvius are not abundant. Gymnotus inaequilabiatus is often misidentified or mistaken for other species, despite the presence of multiple salient external diagnostic characters (Maxime, Albert, 2014Maxime EL, Albert JS. Redescription of the Tuvirão, Gymnotus inaequilabiatus Valenciennes, 1839, using high-resolution x-ray computed tomography. Copeia. 2014; 2014(3):462-72. https://doi.org/10.1643/CI-13-054
https://doi.org/10.1643/CI-13-054...
) that place it in the G. tigre clade rather than the G. carapo clade. Further, G. inaequilabiatus is unique among congeners in its large adult body size, growing to one-meter total length.
We found inconsistent taxonomic identifications when comparing the sequences of the specimens classified here as G. cf. sylvius and G. cf. cuia with the sequences available in GenBank. Three sequences presented better identity with G. inaequilabiatus (clade C), but also grouped with G. paraguensis, G. cf. paraguensis, G. carapo and Gymnotus sp. (Fig. 2). Our sequences of G. cf. cuia presented also 99% of identity with Gymnotus new sp. RS2 that, according to Tagliacollo et al. (2016Tagliacollo VA, Bernt MJ, Craig JM, Oliveira C, Albert JS. Model-based total evidence phylogeny of Neotropical electric knifefishes (Teleostei, Gymnotiformes). Mol Phylogenet Evol. 2016; 95:20-33. https://doi.org/10.1016/j.ympev.2015.11.007
https://doi.org/10.1016/j.ympev.2015.11....
), belongs to G. carapo clade and not to G. tigre clade, which includes the species G. inaequilabiatus and G. paraguensis (Maxime, Albert, 2014Maxime EL, Albert JS. Redescription of the Tuvirão, Gymnotus inaequilabiatus Valenciennes, 1839, using high-resolution x-ray computed tomography. Copeia. 2014; 2014(3):462-72. https://doi.org/10.1643/CI-13-054
https://doi.org/10.1643/CI-13-054...
; Craig et al., 2018Craig JM, Malabarba LR, Crampton WGR, Albert JS. Revision of banded knifefishes of the Gymnotus carapo and G. tigre clades (Gymnotidae Gymnotiformes) from the Southern Neotropics. Zootaxa 2018; 4379(1):47-73. http://dx.doi.org/10.11646/zootaxa.4379.1.3
http://dx.doi.org/10.11646/zootaxa.4379....
). It is possible that the specimens identified as G. inaequilabiatus in previous studies based on Graça, Pavanelli (2007Graça WJ, Pavanelli CS. Peixes da planície de inundação do Alto rio Paraná e áreas adjacentes. Maringá: EDUEM; 2007. ) and Ota et al. (2018Ota RR, Deprá GC, Graça WJ, Pavanelli CS. Peixes da planície de inundação do alto rio Paraná e áreas adjacentes: revised, annotated and updated. Neotrop Ichthyol . 2018; 16(2):e170094. https://doi.10.1590/1982-0224-20170094
https://doi.10.1590/1982-0224-20170094...
) may be actually G. cuia. These discordances were probably caused by misidentification of specimens used in previous studies, once new data from literature, as the recent revisions of Scacchetti et al. (2011Scacchetti PC, Pansonato-Alves JC, Utsunomia R, Oliveira C, Foresti F. Karyotypic diversity in four species of the genus Gymnotus Linnaeus, 1758 (Teleostei, Gymnotiformes, Gymnotidae): Physical mapping of ribosomal genes and telomeric sequences. Comp Cytogenet. 2011; 5(3):223-35. https://doi.org/10.3897/compcytogen.v5i3.1375
https://doi.org/10.3897/compcytogen.v5i3...
), Craig et al. (2017Craig JM, Malabarba LR, Crampton WGR, Albert JS. Revision of banded knifefishes of the Gymnotus carapo and G. tigre clades (Gymnotidae Gymnotiformes) from the Southern Neotropics. Zootaxa 2018; 4379(1):47-73. http://dx.doi.org/10.11646/zootaxa.4379.1.3
http://dx.doi.org/10.11646/zootaxa.4379....
, 2018Drake DAR, Mandrak NE. Ecological risk of live bait fisheries: A new angle on selective fishing. Fisheries 2014; 39(5):201-11. https://doi.org/10.1080/03632415.2014.903835
https://doi.org/10.1080/03632415.2014.90...
) and Utsunomia et al. (2018Utsunomia R, Melo S, Scacchetti PC, Oliveira C, Machado MA, Pieczarka JC. Particular chromosomal distribution of microsatellites in five species of the Genus Gymnotus (Teleostei, Gymnotiformes). Zebrafish . 2018; 15(4):398-403. https://doi.org/10.1089/zeb.2018.1570
https://doi.org/10.1089/zeb.2018.1570...
) are now available to elucidate this complex group.
Despite of these considerations, we demonstrated that at least two different species are fished as live bait in Jacaré-Guaçu River. These two species, preliminarily identified here as G. cf. sylvius and G. cf. cuia, presented 4.7% to 5.6% of genetic divergence (Tab. 1) and they can easily be discriminated by PCR-RFLP method developed in this work. This technique has been proven to efficiently identify different taxa for distinct purposes (Wolf et al., 1999Wolf C, Rentsch J, Hübner P. PCR-RFLP Analysis of mitochondrial DNA: A reliable method for species identification. J Agric Food Chem. 1999; 47(4):1350-55. https://doi.org/10.1021/jf9808426
https://doi.org/10.1021/jf9808426...
; Bieliková et al., 2010Bieliková M, Pangallo D, Turňa J. Polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) as a molecular discrimination tool for raw and heat-treated game and domestic animal meats. J Food Nutr Res. 2010; 49(3):134-39.; Schmidt et al., 2015Schmidt BF, Amorim AF, Hilsdorf AWS. PCR-RFLP analysis to identify four ray species of the genus Dasyatis (Elasmobranchii, Dasyatidae) fished along the southeastern and southern coast of Brazil. Fish Res. 2015; 167:71-74. https://doi.org/10.1016/j.fishres.2014.12.025
https://doi.org/10.1016/j.fishres.2014.1...
). The PCR-RFLP technique developed herein showed that the NlaIII and SacI endonucleases cut patterns clearly distinguished G. pantanal and G. pantherinus species where they occur in sympatry with G. cf. sylvius and G. cf. cuia, as shown in Tabs. 2 - 3. The data demonstrated that the most frequent species was G. cf. sylvius corresponding to 95.2% of the fished specimens. Fernandes-Matioli et al. (2000Fernandes-Matioli FMC, Matioli SR, Almeida-Toledo LF. Species diversity and geographic distribution of Gymnotus (Pisces: Gymnotiformes) by nuclear (GGAC)n microsatellite analysis. Genet Mol Biol. 2000; 23(4): 803-07. http://dx.doi.org/10.1590/S1415-47572000000400016
http://dx.doi.org/10.1590/S1415-47572000...
) also observed Gymnotus populations of different species living in sympatry in some watersheds of Southeastern Brazil and always one of the species occur at a significantly lower frequency. The sequences alignment and analysis of the haplotypes retrieved from GenBank database allowed a carefully examination of polymorphic restriction sites to detect variation among species, excluding sites with intra-specific variation.
The identification of Gymnotus species fished for live bait is extremely important for the management of this fishery resource and as a result to determine proper fisheries regulation. The current fishing legislation in the Paraná River Basin refers to Gymnotus species as G. carapo and determines the minimum capture size of 20 mm (IBAMA, 2009Instituto Brasileiro do Meio Ambiente (IBAMA). Instrução Normativa IBAMA nº 26, de 2 de Setembro de 2009. Brasília: Diário Oficial da União; 2009. Available from: http://www.icmbio.gov.br/cepsul/images/stories/legislacao/Instrucao_normativa/2009/in_ibama_26_2009_normaspescabaciahidrografica_rioparana_rev_in_30_2005.pdf
http://www.icmbio.gov.br/cepsul/images/s...
). Thus, the present outcomes show the need for legislation change to include other Gymnotus being caught as live bait. The minimum capture size depends on the captured species, since each species has its reproduction cycle. Sousa et al. (2017Sousa TP, Marques DKS, Vitorino CA, Faria KC, Braga GSF, Ferreira DC, Venere PC. Cytogenetic and molecular data support the occurrence of three Gymnotus Species (Gymnotiformes: Gymnotidae) used as live bait in Corumbá, Brazil: implications for conservation and management of professional fishing. Zebrafish. 2017; 14(2):177-86. https://doi.org/10.1089/zeb.2016.1356
https://doi.org/10.1089/zeb.2016.1356...
) also suggested a revision of capture legislation in the Pantanal of Mato Grosso do Sul, because it recognizes two Gymnotus species. However, these authors found three species of Gymnotus commercialized as live bait in the region.
The use of live bait has been concerning worldwide due to risks of introduction of invasive alien species by sport anglers and the ecological impact on the native biodiversity once this live bait species become established (Sá et al., 2017Sá E, Costa PF, Fonseca LC, Alves AS, Castro N, Cabral SS, Chainho P, Canning-Clode J, Melo P, Pombo AM, Costa JL. Trade of live bait in Portugal and risks of introduction of nonindigenous species associated to importation. Ocean Coast Manage. 2017; 146:121-28. https://doi.org/10.1016/j.ocecoaman.2017.06.016
https://doi.org/10.1016/j.ocecoaman.2017...
). At the same time, even local fish species being indiscriminately used as live bait can impact the long-term survival of local species population. Species identification and management through legal monitoring is central to maintain sustainable fishing by local communities. The molecular surveillance tool developed herein allows the implementation of a rapid and low-cost diagnostic system by PCR-RFLP to identify Gymnotus species in the sampling fishing area of this study, which can be applied to yield seasonal Gymnotus fishing data and support a reliable fishing regulation to use this fisheries resources sustainably.
Acknowledgments
We gratefully acknowledge the fishing community of Fazenda São Giacomo and the Jacaré-Guaçu River, Ibitinga, SP in contributing to this work. The team of the Laboratório de Genética de Organismos Aquáticos e Aquicultura (Lagoaa) at the Universidade de Mogi das Cruzes (UMC) and Fundação de Apoio a Ensino e Pesquisa (FAEP) for the support and assistance during the laboratory analyzes. Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) by the scholarship of Dr. Alexandre W. S. Hilsdorf. Dr. Luiz A. W. Peixoto and Dr. Michel D. Gianeti from MZUSP, for help in identifying some specimens. Dr. Paula M. de Castro from Instituto de Pesca (IP), SP, for assisting in the planning of this work. Messrs. Luiz Evangelista and Sérgio Silva from IP, for assisting in field surveys.
References
- Albert JS. Species diversity and phylogenetic systematics of American knifefishes (Gymnotiformes, Teleostei). Misc Publ Mus Zool Univ Mich. 2001; 190:1-127.
- Albert JS, Crampton WGR. Seven new species of the Neotropical electric fish Gymnotus (Teleostei, Gymnotiformes) with a redescription of G. carapo (Linnaeus). Zootaxa. 2003; 287(1):1-54. http://dx.doi.org/10.11646/zootaxa.287.1.1
» http://dx.doi.org/10.11646/zootaxa.287.1.1 - Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997; 25(22):4692-93. https://doi.org/10.1093/nar/25.22.4692
» https://doi.org/10.1093/nar/25.22.4692 - Alves-Gomes JA, Ortí G, Haygood M, Heiligenberg W, Meyer A. Phylogenetic analysis of the South American electric fishes (Order Gymnotiformes) and the evolution of their electrogenic system: A synthesis based on morphology, electrophysiology, and mitochondrial sequence data. Mol Biol Evol. 1995; 12(2):298-318. https://doi.org/10.1093/oxfordjournals.molbev.a040204
» https://doi.org/10.1093/oxfordjournals.molbev.a040204 - Bieliková M, Pangallo D, Turňa J. Polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) as a molecular discrimination tool for raw and heat-treated game and domestic animal meats. J Food Nutr Res. 2010; 49(3):134-39.
- Craig JM, Crampton WGR, Albert JS. Revision of the polytypic electric fish Gymnotus carapo (Gymnotiformes, Teleostei), with descriptions of seven species. Zootaxa . 2017; 4318(3):401-38. http://dx.doi.org/10.11646/zootaxa.4318.3.1
» http://dx.doi.org/10.11646/zootaxa.4318.3.1 - Craig JM, Malabarba LR, Crampton WGR, Albert JS. Revision of banded knifefishes of the Gymnotus carapo and G. tigre clades (Gymnotidae Gymnotiformes) from the Southern Neotropics. Zootaxa 2018; 4379(1):47-73. http://dx.doi.org/10.11646/zootaxa.4379.1.3
» http://dx.doi.org/10.11646/zootaxa.4379.1.3 - Drake DAR, Mandrak NE. Ecological risk of live bait fisheries: A new angle on selective fishing. Fisheries 2014; 39(5):201-11. https://doi.org/10.1080/03632415.2014.903835
» https://doi.org/10.1080/03632415.2014.903835 - Fernandes-Matioli FMC, Matioli SR, Almeida-Toledo LF. Species diversity and geographic distribution of Gymnotus (Pisces: Gymnotiformes) by nuclear (GGAC)n microsatellite analysis. Genet Mol Biol. 2000; 23(4): 803-07. http://dx.doi.org/10.1590/S1415-47572000000400016
» http://dx.doi.org/10.1590/S1415-47572000000400016 - Fernandes FMC, Albert JS, Daniel-Silva MFZ, Lopes CE, Crampton WGR, Almeida-Toledo LF. A new Gymnotus (Teleostei: Gymnotiformes: Gymnotidae) from the Pantanal Matogrossense of Brazil and adjacent drainages: Continued documentation of a cryptic fauna. Zootaxa . 2005; 933(1):1-14. http://dx.doi.org/10.11646/zootaxa.933.1.1
» http://dx.doi.org/10.11646/zootaxa.933.1.1 - Food and Agriculture Organization (FAO). Fish identification tools for biodiversity and fisheries assessments: Review and guidance for decision-makers. Rome: Food and Agriculture Organization of the United Nations [Internet]. Rome; 2013. Available from: http://www.fao.org/docrep/019/i3354e/i3354e.pdf
» http://www.fao.org/docrep/019/i3354e/i3354e.pdf - Graça WJ, Pavanelli CS. Peixes da planície de inundação do Alto rio Paraná e áreas adjacentes. Maringá: EDUEM; 2007.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc B. 2003; 270(1512):313-21. https://doi.org/10.1098/rspb.2002.2218
» https://doi.org/10.1098/rspb.2002.2218 - Hilsdorf AWS, Hallerman EM. Genetic resources of Neotropical fishes. Cham, Switzerland: Springer International Publishing AG; 2017.
- Instituto Brasileiro do Meio Ambiente (IBAMA). Instrução Normativa IBAMA nº 26, de 2 de Setembro de 2009. Brasília: Diário Oficial da União; 2009. Available from: http://www.icmbio.gov.br/cepsul/images/stories/legislacao/Instrucao_normativa/2009/in_ibama_26_2009_normaspescabaciahidrografica_rioparana_rev_in_30_2005.pdf
» http://www.icmbio.gov.br/cepsul/images/stories/legislacao/Instrucao_normativa/2009/in_ibama_26_2009_normaspescabaciahidrografica_rioparana_rev_in_30_2005.pdf - Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16(2):111-20.
- Margarido VP, Bellafronte E, Moreira-Filho O. Cytogenetic analysis of three sympatric Gymnotus (Gymnotiformes, Gymnotidae) species verifies invasive species in the Upper Paraná River basin, Brazil. J Fish Biol. 2007; 70(sb):155-64. https://doi.org/10.1111/j.1095-8649.2007.01365.x
» https://doi.org/10.1111/j.1095-8649.2007.01365.x - Maxime EL, Albert JS. A new species of Gymnotus (Gymnotiformes: Gymnotidae) from the Fitzcarrald Arch of southeastern Peru. Neotrop Ichthyol. 2009; 7(4):579-85. http://dx.doi.org/10.1590/S1679-62252009000400004
» http://dx.doi.org/10.1590/S1679-62252009000400004 - Maxime EL, Albert JS. Redescription of the Tuvirão, Gymnotus inaequilabiatus Valenciennes, 1839, using high-resolution x-ray computed tomography. Copeia. 2014; 2014(3):462-72. https://doi.org/10.1643/CI-13-054
» https://doi.org/10.1643/CI-13-054 - Milhomem SSR, Crampton WGR, Pieczarka JC, Shetka GH, Silva DS, Nagamachi CY. Gymnotus capanema, a new species of electric knife fish (Gymnotiformes, Gymnotidae) from eastern Amazonia, with comments on an unusual karyotype. J Fish Biol . 2012; 80(4):802-15. https://doi.org/10.1111/j.1095-8649.2012.03219.x
» https://doi.org/10.1111/j.1095-8649.2012.03219.x - Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics 2008; 24(16):1757-64. https://doi.org/10.1093/bioinformatics/btn322
» https://doi.org/10.1093/bioinformatics/btn322 - Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000.
- Ogden R. Fisheries forensics: The use of DNA tools for improving compliance, traceability and enforcement in the fishing industry. Fish Fish. 2008; 9(4):462-72. https://doi.org/10.1111/j.1467-2979.2008.00305.x
» https://doi.org/10.1111/j.1467-2979.2008.00305.x - Ota RR, Deprá GC, Graça WJ, Pavanelli CS. Peixes da planície de inundação do alto rio Paraná e áreas adjacentes: revised, annotated and updated. Neotrop Ichthyol . 2018; 16(2):e170094. https://doi.10.1590/1982-0224-20170094
» https://doi.10.1590/1982-0224-20170094 - Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003; 19(18):2496-97. https://doi.org/10.1093/bioinformatics/btg359
» https://doi.org/10.1093/bioinformatics/btg359 - Sá E, Costa PF, Fonseca LC, Alves AS, Castro N, Cabral SS, Chainho P, Canning-Clode J, Melo P, Pombo AM, Costa JL. Trade of live bait in Portugal and risks of introduction of nonindigenous species associated to importation. Ocean Coast Manage. 2017; 146:121-28. https://doi.org/10.1016/j.ocecoaman.2017.06.016
» https://doi.org/10.1016/j.ocecoaman.2017.06.016 - Scacchetti PC, Pansonato-Alves JC, Utsunomia R, Oliveira C, Foresti F. Karyotypic diversity in four species of the genus Gymnotus Linnaeus, 1758 (Teleostei, Gymnotiformes, Gymnotidae): Physical mapping of ribosomal genes and telomeric sequences. Comp Cytogenet. 2011; 5(3):223-35. https://doi.org/10.3897/compcytogen.v5i3.1375
» https://doi.org/10.3897/compcytogen.v5i3.1375 - Schmidt BF, Amorim AF, Hilsdorf AWS. PCR-RFLP analysis to identify four ray species of the genus Dasyatis (Elasmobranchii, Dasyatidae) fished along the southeastern and southern coast of Brazil. Fish Res. 2015; 167:71-74. https://doi.org/10.1016/j.fishres.2014.12.025
» https://doi.org/10.1016/j.fishres.2014.12.025 - Sousa TP, Marques DKS, Vitorino CA, Faria KC, Braga GSF, Ferreira DC, Venere PC. Cytogenetic and molecular data support the occurrence of three Gymnotus Species (Gymnotiformes: Gymnotidae) used as live bait in Corumbá, Brazil: implications for conservation and management of professional fishing. Zebrafish. 2017; 14(2):177-86. https://doi.org/10.1089/zeb.2016.1356
» https://doi.org/10.1089/zeb.2016.1356 - Tagliacollo VA, Bernt MJ, Craig JM, Oliveira C, Albert JS. Model-based total evidence phylogeny of Neotropical electric knifefishes (Teleostei, Gymnotiformes). Mol Phylogenet Evol. 2016; 95:20-33. https://doi.org/10.1016/j.ympev.2015.11.007
» https://doi.org/10.1016/j.ympev.2015.11.007 - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol . 2014; 30(12):2725-29. https://doi.org/10.1093/molbev/mst197
» https://doi.org/10.1093/molbev/mst197 - Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res . 1994; 22(22):4673-80. https://doi.org/10.1093/nar/22.22.4673
» https://doi.org/10.1093/nar/22.22.4673 - Utsunomia R, Melo S, Scacchetti PC, Oliveira C, Machado MA, Pieczarka JC. Particular chromosomal distribution of microsatellites in five species of the Genus Gymnotus (Teleostei, Gymnotiformes). Zebrafish . 2018; 15(4):398-403. https://doi.org/10.1089/zeb.2018.1570
» https://doi.org/10.1089/zeb.2018.1570 - Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res . 2003; 31(13):3688-91. https://doi.org/10.1093/nar/gkg526
» https://doi.org/10.1093/nar/gkg526 - Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philos Trans R Soc London B. 2005; 360(1462):1847-57. https://doi.org/10.1098/rstb.2005.1716
» https://doi.org/10.1098/rstb.2005.1716 - Wolf C, Rentsch J, Hübner P. PCR-RFLP Analysis of mitochondrial DNA: A reliable method for species identification. J Agric Food Chem. 1999; 47(4):1350-55. https://doi.org/10.1021/jf9808426
» https://doi.org/10.1021/jf9808426 - Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000; 7(1-2):203-14. https://doi.org/10.1089/10665270050081478
Edited by
Publication Dates
-
Publication in this collection
25 Nov 2019 -
Date of issue
2019
History
-
Received
22 July 2019 -
Accepted
17 Oct 2019