Abstract
Anaplasma marginale is a vector-borne pathogen that causes a disease known as anaplasmosis. No sequenced genomes of Brazilian strains are yet available. The aim of this work was to compare whole genomes of Brazilian strains of A. marginale (Palmeira and Jaboticabal) with genomes of strains from other regions (USA and Australia strains). Genome sequencing of Brazilian strains was performed by means of next-generation sequencing. Reads were mapped using the genome of the Florida strain of A. marginale as a reference sequence. Single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs) were identified. The data showed that two Brazilian strains grouped together in one particular clade, which grouped in a larger American group together with North American strains. Moreover, some important differences in surface proteins between the two Brazilian isolates can be discerned. These results shed light on the evolutionary history of A. marginale and provide the first genome information on South American isolates. Assessing the genome sequences of strains from different regions is essential for increasing knowledge of the pan-genome of this bacteria.
Keywords:
Cattle tick fever; bovine anaplasmosis; Rickettsiales; genome; next-generation sequencing
Resumo
Anaplasma marginale é um patógeno transmitido por vetores que causam uma doença conhecida como anaplasmose. Até a presente data, não há genomas sequenciados de cepas brasileiras. O objetivo deste estudo foi comparar o genoma completo das cepas brasileiras de A. marginale (Palmeira e Jaboticabal) com os genomas de cepas de outras regiões (cepas dos EUA e Austrália). As sequências dos genomas das cepas brasileiras foram obtidas mediante sequenciamento de nova geração. As “reads” foram mapeadas usando-se como referência o genoma de A. marginale da cepa Florida. Foram identificados polimorfismos de nucleotídeo único (SNPs) e analisadas inserções/deleções (INDELs). As duas linhagens brasileiras se agruparam em um clado particular que, por sua vez, agrupou-se em um grupo maior junto com as linhagens norte-americanas. Além disso, foram identificadas diferenças significativas nas proteínas de superfície entre os dois isolados brasileiros. Esses resultados lançam luz sobre a história evolutiva de A. marginale e fornecem as primeiras informações de genomas de isolados sul-americanos. Avaliar as sequências de genomas de cepas de diferentes regiões é essencial para aumentar o conhecimento do pan-genoma dessa bactéria.
Palavras-chave:
Tristeza Parasitária Bovina; anaplasmose bovina; Rickettsiales; genoma; sequenciamento de nova geração
Introduction
Anaplasma marginale is an obligate intracellular, vector-borne pathogen belonging to the order Rickettsiales, family Anaplasmataceae, which causes a disease known as anaplasmosis. Bovine anaplasmosis occurs in tropical and subtropical areas throughout the world, and this disease is a major constraint on cattle production in several countries (Kocan et al., 2010Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol 2010; 167(2-4): 95-107. http://dx.doi.org/10.1016/j.vetpar.2009.09.012. PMid:19811876.
http://dx.doi.org/10.1016/j.vetpar.2009....
, 2015Kocan KM, de la Fuente J, Cabezas-Cruz A. The genus Anaplasma: new challenges after reclassification. Rev Sci Tech 2015; 34(2): 577-586. http://dx.doi.org/10.20506/rst.34.2.2381. PMid:26601458.
http://dx.doi.org/10.20506/rst.34.2.2381...
). Previous research reported differences in virulence among A. marginale strains (da Silva et al., 2016da Silva JB, André MR, Machado RZ. Low genetic diversity of Anaplasma marginale in calves in an endemic area for bovine anaplasmosis in the state of São Paulo, Brazil. Ticks Tick Borne Dis 2016; 7(1): 20-25. http://dx.doi.org/10.1016/j.ttbdis.2015.07.018. PMid:26318264.
http://dx.doi.org/10.1016/j.ttbdis.2015....
; Machado et al., 2015Machado RZ, Silva JB, André MR, Gonçalves LR, Matos CA, Obregón D. Outbreak of anaplasmosis associated with the presence of different Anaplasma marginale strains in dairy cattle in the states of São Paulo and Goiás, Brazil. Rev Bras Parasitol Vet 2015; 24(4): 438-446. http://dx.doi.org/10.1590/S1984-29612015078. PMid:26648009.
http://dx.doi.org/10.1590/S1984-29612015...
; Silva et al., 2015Silva JB, Gonçalves LR, Varani AM, André MR, Machado RZ. Genetic diversity and molecular phylogeny of Anaplasma marginale studied longitudinally under natural transmission conditions in Rio de Janeiro, Brazil. Ticks Tick Borne Dis 2015; 6(4): 499-507. http://dx.doi.org/10.1016/j.ttbdis.2015.04.002. PMid:25985719.
http://dx.doi.org/10.1016/j.ttbdis.2015....
), which indicates that there is a need for better understanding of the genetic mechanisms involved in A. marginale infection and the pathogenesis of anaplasmosis.
Availability of genome sequences obtained through next-generation sequencing (NGS) has revolutionized the field of infectious disease research. The plethora of data not only has enabled advances in fundamental biology, thereby helping to unveil the pathogenesis of infections and the genomic evolution of microorganisms, but also has contributed to advances in clinical microbiology (Fournier et al., 2014Fournier PE, Dubourg G, Raoult D. Clinical detection and characterization of bacterial pathogens in the genomics era. Genome Med 2014; 6(11): 114. http://dx.doi.org/10.1186/s13073-014-0114-2. PMid:25593594.
http://dx.doi.org/10.1186/s13073-014-011...
).
Members of the order Rickettsiales are small, obligate intracellular bacteria (Dumler et al., 2001Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol 2001; 51(Pt6): 2145-2165. http://dx.doi.org/10.1099/00207713-51-6-2145. PMid:11760958.
http://dx.doi.org/10.1099/00207713-51-6-...
) that typically have small genomes, which is attributed to reductive evolution following long-term intracellular parasitism (Andersson et al., 1998Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998; 396(6707): 133-140. http://dx.doi.org/10.1038/24094. PMid:9823893.
http://dx.doi.org/10.1038/24094...
; Brayton et al., 2005Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 2005; 102(3): 844-849. http://dx.doi.org/10.1073/pnas.0406656102. PMid:15618402.
http://dx.doi.org/10.1073/pnas.040665610...
; Ogata et al., 2001Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 2001; 293(5537): 2093-2098. http://dx.doi.org/10.1126/science.1061471. PMid:11557893.
http://dx.doi.org/10.1126/science.106147...
; Wu et al., 2004Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2004; 2(3): e69. http://dx.doi.org/10.1371/journal.pbio.0020069. PMid:15024419.
http://dx.doi.org/10.1371/journal.pbio.0...
). The first genome of A. marginale was published in 2005 and showed that the surface coat is composed by many surface proteins, which includes, in addition to several outer membrane proteins, two gene families encoding immunodominant proteins, the major surface protein 1 (msp1) and major surface protein 2 (msp2) superfamilies. Out of the 949 annotated coding sequences, just 62 are predicted to be outer membrane proteins and, of these, 49 belong to one of these two superfamilies.
The genome contains unusual functional pseudogenes that belong to the msp2 superfamily and are involved in surface coat antigenic variation. This differs from pseudogenes that are described as byproducts of reductive evolution in other members of Rickettsiales (Brayton et al., 2005Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 2005; 102(3): 844-849. http://dx.doi.org/10.1073/pnas.0406656102. PMid:15618402.
http://dx.doi.org/10.1073/pnas.040665610...
). Comparisons within North American strains have revealed that A. marginale has a closed-core genome with a few highly plastic regions. These include the msp2 and msp3 genes and the appendage-associated protein (aaap) locus (Dark et al., 2009Dark MJ, Herndon DR, Kappmeyer LS, Gonzales MP, Nordeen E, Palmer GH, et al. Conservation in the face of diversity: multistrain analysis of an intracellular bacterium. BMC Genomics 2009; 10(1): 16. http://dx.doi.org/10.1186/1471-2164-10-16. PMid:19134224.
http://dx.doi.org/10.1186/1471-2164-10-1...
).
Analysis on multiple A. marginale strains has suggested that intracellular bacteria have more variable single nucleotide polymorphism (SNP) retention rates than previously reported. Moreover, they may have closed-core genomes that have developed in response to the host organism environment and/or reductive evolution (Dark et al., 2009Dark MJ, Herndon DR, Kappmeyer LS, Gonzales MP, Nordeen E, Palmer GH, et al. Conservation in the face of diversity: multistrain analysis of an intracellular bacterium. BMC Genomics 2009; 10(1): 16. http://dx.doi.org/10.1186/1471-2164-10-16. PMid:19134224.
http://dx.doi.org/10.1186/1471-2164-10-1...
). Recently, phylogenetic analyses on two Australian strains revealed that they presented a marked evolutionary distance from North American strains. Single nucleotide polymorphism analysis showed strikingly reduced genetic diversity among the Australian strains, with the smallest number of SNPs detected between two A. marginale strains (Pierlé et al., 2014Pierlé S, Imaz-Rosshandler I, Kerudin A, Sambono J, Lew-Tabor A, Rolls P, et al. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens 2014; 3(1): 57-72. http://dx.doi.org/10.3390/pathogens3010057. PMid:25364572.
http://dx.doi.org/10.3390/pathogens30100...
).
No South American genomes for A. marginale have yet become available. In fact, genomic information is only available for strains from North America and Australia. Regarding Brazilian A. marginale strains, analyses on genetic diversity have been based on molecular markers such as the msp4 and msp1α genes (Baêta et al., 2015Baêta BA, Ribeiro CCDU, Teixeira RC, Cabezas-Cruz A, Passos LMF, Zweygarth E, et al. Characterization of two strains of Anaplasma marginale isolated from cattle in Rio de Janeiro, Brazil, after propagation in tick cell culture. Ticks Tick Borne Dis 2015; 6(2): 141-145. http://dx.doi.org/10.1016/j.ttbdis.2014.11.003. PMid:25468764.
http://dx.doi.org/10.1016/j.ttbdis.2014....
; de la Fuente et al., 2002de la Fuente J, Van Den Bussche RA, Garcia-Garcia JC, Rodríguez SD, García MA, Guglielmone AA, et al. Phylogeography of New World isolates of Anaplasma marginale based on major surface protein sequences. Vet Microbiol 2002; 88(3): 275-285. http://dx.doi.org/10.1016/S0378-1135(02)00122-0. PMid:12151201.
http://dx.doi.org/10.1016/S0378-1135(02)...
, 2003de la Fuente J, Van Den Bussche RA, Prado TM, Kocan KM. Anaplasma marginale msp1α genotypes evolved under positive selection pressure but are not markers for geographic isolates. J Clin Microbiol 2003; 41(4): 1609-1616. http://dx.doi.org/10.1128/JCM.41.4.1609-1616.2003. PMid:12682152.
http://dx.doi.org/10.1128/JCM.41.4.1609-...
, 2004de La Fuente J, Passos LMF, Van Den Bussche RA, Ribeiro MFB, Facury-Filho EJ, Kocan KM. Genetic diversity and molecular phylogeny of Anaplasma marginale isolates from Minas Gerais, Brazil. Vet Parasitol 2004; 121(3-4): 307-316. http://dx.doi.org/10.1016/j.vetpar.2004.02.021. PMid:15135871.
http://dx.doi.org/10.1016/j.vetpar.2004....
; Pohl et al., 2013Pohl AE, Cabezas-Cruz A, Ribeiro MFB, Silveira JAG, Silaghi C, Pfister K, et al. Detection of genetic diversity of Anaplasma marginale isolates in Minas Gerais, Brazil. Rev Bras Parasitol Vet 2013; 22(1): 129-135. http://dx.doi.org/10.1590/S1984-29612013000100024. PMid:24252959.
http://dx.doi.org/10.1590/S1984-29612013...
; Silva et al., 2014aSilva JB, Cabezas-Cruz A, Fonseca AH, Barbosa JD, de la Fuente J. Infection of water buffalo in Rio de Janeiro Brazil with Anaplasma marginale strains also reported in cattle. Vet Parasitol 2014a; 205(3-4): 730-734. http://dx.doi.org/10.1016/j.vetpar.2014.09.009. PMid:25260335.
http://dx.doi.org/10.1016/j.vetpar.2014....
,bSilva JB, Fonseca AH, Barbosa JD, Cabezas-Cruz A, de la Fuente J. Low genetic diversity associated with low prevalence of Anaplasma marginale in water buffaloes in Marajó Island, Brazil. Ticks Tick Borne Dis 2014b; 5(6): 801-804. http://dx.doi.org/10.1016/j.ttbdis.2014.06.003. PMid:25108778.
http://dx.doi.org/10.1016/j.ttbdis.2014....
; Vidotto et al., 2006Vidotto MC, Kano SF, Gregori F, Headley SA, Vidotto O. Phylogenetic analysis of Anaplasma marginale strains from Paraná State, Brazil, using the msp1α and msp4 genes. J Vet Med B Infect Dis Vet Public Health 2006; 53(9): 404-411. http://dx.doi.org/10.1111/j.1439-0450.2006.00984.x. PMid:17062116.
http://dx.doi.org/10.1111/j.1439-0450.20...
). Information about A. marginale in southern Brazil is even scarcer. It is noteworthy that cattle tick fever (a popular expression that in Brazil usually refers both to cattle babesiosis and/or anaplamosis) is the most significant cause of cattle death in this region. Indeed, the climate, vector epidemiology and cattle breeding systems of this region are markedly different from those of the rest of the country.
In this context, the aim of the present study was to characterize and analyze the first genome sequences of Brazilian strains of A. marginale, with comparison of two strains from different Brazilian regions (south and southeast). In addition, the sequences obtained from the Brazilian strains were compared with the genomes of strains from other countries in order to contribute to global and local knowledge of bovine anaplasmosis.
Materials and Methods
Anaplasma strains
Two strains of A. marginale were evaluated in this work. The A. marginale strain Palmeira was isolated in the early 2000s, from Holstein dairy cattle that showed clinical anaplasmosis from the municipality of Palmeira das Missões, state of Rio Grande do Sul, southern Brazil (27° 54' 09” S; 53° 18' 26” W). The Jaboticabal strain was isolated from cattle in the municipality of Jaboticabal, São Paulo state, southeast Brazil (21° 15' 18” S; 48° 19' 20” W). After isolation, both strains were cryopreserved with 10% dimethyl sulfoxide (DMSO) in liquid nitrogen. The Palmeira strain was isolated from a region where there had been massive use of a heterologous live vaccine based on Anaplasma centrale. Indeed, after clinical cases of anaplasmosis caused by the Palmeira strain had been identified, occurrences of novel anaplasmosis outbreaks on the ranch were avoided by vaccinating the cattle with A. centrale. On the other hand, the Jaboticabal strain was originally isolated from a region where the A. centrale live vaccine is rarely used. There is no evidence that any of two studied strains is tick transmissible. It is important to note that in Brazil, the only species with economic and epidemiological importance as tick of cattle is Rhipicephalus microplus. And, in this sense, to date, there is no clear/unquestionable evidence in the literature on A. marginale transovarial transmission in R. microplus ticks (Esteves et al., 2015Esteves E, Pohl PC, Klafke GM, Reck J, Fogaça AC, Martins JR, et al. Low temperature affects cattle tick reproduction but does not lead to transovarial transmission of Anaplasma marginale. Vet Parasitol 2015; 214(3-4): 322-326. http://dx.doi.org/10.1016/j.vetpar.2015.07.010. PMid:26255094.
http://dx.doi.org/10.1016/j.vetpar.2015....
).
DNA isolation and library construction
DNA was isolated from cryopreserved blood using the PureLink genomic DNA mini-kit (Invitrogen™, Carlsbad, CA, USA), following the manufacturer's recommendations. The DNA samples were quantified using NanoDrop (Thermo Scientific, Waltham, MA, USA) and Qubit (Life Technologies™, Carlsbad, CA, USA), and DNA integrity was checked by means of agarose gel electrophoresis. For library synthesis, the DNA was cleaved using transposase (Nextera DNA sample preparation kit; Illumina®, San Diego, CA, USA) and purified using a commercial kit (Qiaquick PCR purification kit; Qiagen, Hilden, Germany). After that, the samples were indexed and ligated to Illumina-specific adapters, which, respectively, allow post-sequencing sample identification and linkage to the flow cell (Nextera index kit; Illumina®, San Diego, CA, USA). The libraries were amplified by means of the polymerase chain reaction (PCR) and then purified (AMPure XP; Beckman Coulter Inc, Brea, CA, USA) and quantified by means of real-time PCR (RT-PCR), using SYBR green and PhiX as the control (PhiX control kit v3; Illumina®, San Diego, CA, USA).
Genome sequencing, assembly and annotation
Genome sequencing was performed in the MiSeq desktop sequencer (Illumina®, San Diego, CA, USA) using the Miseq reagent kit v2 and v3. Raw fastq files were mapped using the Geneious 9.1.2 software (Biomatters Ltd., Auckland, New Zealand) (Kearse et al., 2012Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28(12): 1647-1649. http://dx.doi.org/10.1093/bioinformatics/bts199. PMid:22543367.
http://dx.doi.org/10.1093/bioinformatics...
; Ripma et al., 2014Ripma LA, Simpson MG, Hasenstab-Lehman KE. Geneious! Simplified genome skimming methods for phylogenetic systematic studies: a case study in Oreocarya (Boraginaceae). Appl Plant Sci 2014; 2(12): 1400062. http://dx.doi.org/10.3732/apps.1400062. PMid:25506521.
http://dx.doi.org/10.3732/apps.1400062...
). The A. marginale Florida strain genome (USA origin) (GenBank reference: NC012026) was used as a reference for assembling the genomes of the Palmeira and Jaboticabal strains. The reads were assembled into a single pseudochromosome against the genome of the reference strain. Coding sequence (CDS) annotation and circular genome maps were made using the Geneious software. Only gene sequences of similarity higher than 95% in relation to the St. Maries reference strain were annotated.
Sequence analyses
In order to allow identification of SNPs and insertions/deletions (INDELs), the A. marginale Palmeira and Jaboticabal strain sequences were aligned with the genome of the Florida strain (Reference) using the LASTZ alignment tool, version 1.02.00 (Harris, 2007Harris RS. Improved pairwise alignment of genomic DNA. State College: Penn State University Press; 2007.; Schwartz et al., 2003Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, et al. Human-mouse alignments with BLASTZ. Genome Res 2003; 13(1): 103-107. http://dx.doi.org/10.1101/gr.809403. PMid:12529312.
http://dx.doi.org/10.1101/gr.809403...
), from the Geneious software. SNPs were identified among selected sequences using the Find Variations/SNPs tool from the Geneious software. For the A. marginale Palmeira and Jaboticabal strains, the SNP calling procedure was performed independently for each reference strain. The data generated from the SNP analyses were exported to a Microsoft Excel spreadsheet (Microsoft, Redmond, WA, USA) to build a Venn diagram. The INDEL analyses were also performed using the Find Variations tool from the Geneious software, to search for INDELs. For phylogenomic analysis, genome sequences were analyzed by means of the maximum likelihood algorithm, using the PhyML method with the Seaview 4 software (Gouy et al., 2010Gouy M, Guindon S, Gascuel O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol Biol Evol 2010; 27(2): 221-224. http://dx.doi.org/10.1093/molbev/msp259. PMid:19854763.
http://dx.doi.org/10.1093/molbev/msp259...
).
Results and Discussion
High genome coverages were obtained for the A. marginale Palmeira and Jaboticabal strains, of 28.1 and 49.6-fold, respectively. A total of 317,067 paired-end reads were obtained, and the fragments ranged from 35 to 301 bp (49.8% G/C content) for the Jaboticabal strain. For the Palmeira strain, 141,491 paired-end reads were obtained, and the fragments ranged from 35 to 251 bp (49.8% G/C content). The Palmeira and Jaboticabal strains were aligned using the genome sequence of the Florida strain to produce single contiguous pseudochromosomes of 1,195,100 and 1,195,221 nucleotides, respectively (Figure 1). The annotation of these genomes resulted in a total of 894 CDs in the Palmeira strain and 888 CDs in the Jaboticabal strain. FASTQ files containing raw sequences and sequence qualities of the Palmeira and Jaboticabal strains were deposited at the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under the accession number SRP091646, and Bio Project Accession number PRJNA348690. Genome sequences of the Palmeira and Jaboticabal strains were deposited at the genome databank of NCBI under the accession numbers CP023730.1 and CP023731.1, respectively.
Circular display of the annotated chromosomes of the Palmeira (A) and Jaboticabal (B) strains of A. marginale. The inner circles show the GC (blue) and AT (green) content of the genome. Arrows in the outer circle represent genes (green), coding sequences (CDS) (yellow), transfer RNA (tRNA) (pink), ribosomal RNA (rRNA) (red) and sequence-tagged sites (Sts) (white).
Pairwise comparison of the genomic sequences revealed symmetric identity of 98.5% between the two Brazilian strains, using NCBI genome neighbor reports. Table 1 shows a summary of symmetric identity among Brazilian isolates and some available A. marginale genomes. As expected, the Brazilian strains showed higher identity to each other than to isolates from other countries. Phylogenomic analysis grouped the Palmeira and Jaboticabal strains in one particular clade near to the North American Florida strain, and to the North American St. Maries strain. On the other hand, the Brazilian strains were separate from the Australian strains (Gypsy Plains and Dawn) (Figure 2).
Symmetric identity (%) and gapped identity (%) of A. marginale strains from Brazil (Jaboticabal and Palmeira), the United States (Florida and St. Maries) and Australia (Dawn and Gypsy Plains).
Phylogenomic analysis. Genome sequences of the two Brazilian strains (Palmeira and Jaboticabal) of Anaplasma marginale provided in this study were analyzed together with available genomes from the United States (St. Maries and Florida) and Australia (Gypsy Plains and Dawn). For the phylogenomic tree, genome sequences were analyzed by means of the maximum likelihood algorithm using the PhyML method in the Seaview 4 software. Note that three clades were evident on the phylogenomic tree: the Brazilian strains grouped closer to the United States strains than to the Australian ones. The Anaplasma centrale strain Israel was used as an outgroup.
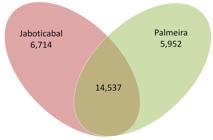
Whole-genome comparisons of A. marginale strains identified 27,203 SNPs in Palmeira and/or Jaboticabal, in comparison with the Florida strain. A total of 5,952 SNPs were unique to the Palmeira strain, while 6,714 SNPs were unique to the Jaboticabal strain, and 14,537 SNPs were common to the two Brazilian strains (see Venn diagram in Figure 3). In comparing the Dawn and Gypsy Plains strains from Australia with the North American St. Maries strain, Pierlé et al. (2014)Pierlé S, Imaz-Rosshandler I, Kerudin A, Sambono J, Lew-Tabor A, Rolls P, et al. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens 2014; 3(1): 57-72. http://dx.doi.org/10.3390/pathogens3010057. PMid:25364572.
http://dx.doi.org/10.3390/pathogens30100...
found 9,813 SNPs that were common to the two Australian strains, while only 97 and 98 of these SNPs were unique to Gypsy Plains and Dawn, respectively. Single nucleotide polymorphism comparison of the Florida strain with four other strains from North America (Puerto Rico, Virginia, Mississippi and St. Maries) revealed the presence of a total of 20,028 sites with SNPs in at least one of the strains; there were 9,609 SNPs in common between the Florida and St. Maries strains, comprising 0.8% of the larger Florida genome (Dark et al., 2009Dark MJ, Herndon DR, Kappmeyer LS, Gonzales MP, Nordeen E, Palmer GH, et al. Conservation in the face of diversity: multistrain analysis of an intracellular bacterium. BMC Genomics 2009; 10(1): 16. http://dx.doi.org/10.1186/1471-2164-10-16. PMid:19134224.
http://dx.doi.org/10.1186/1471-2164-10-1...
).
Single nucleotide polymorphisms (SNPs) identified in the Jaboticabal and Palmeira strains, compared with the Florida strain of A. marginale. The Venn diagram shows the SNPs found in the Jaboticabal and Palmeira strains of A. marginale, using the Florida strain as a reference. A total of 27,203 SNPs (sum of all elements of the Venn diagram) were identified.
Regarding INDEL analysis, comparison of the genomes of the Brazilian strains showed the presence of 3,043 INDELs, encompassing 11,899 bp, considering Florida as the reference strain. There was just one large INDEL region in the Palmeira and Jaboticabal strains, compared with the reference strain. Deletions of 141 bp and 19 bp in the omp9 gene sequence were found in the Palmeira and Jaboticabal strains, respectively.
A large deletion in the omp9 gene had previously been reported for the Australian Dawn and Gypsy Plains strains (Pierlé et al., 2014Pierlé S, Imaz-Rosshandler I, Kerudin A, Sambono J, Lew-Tabor A, Rolls P, et al. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens 2014; 3(1): 57-72. http://dx.doi.org/10.3390/pathogens3010057. PMid:25364572.
http://dx.doi.org/10.3390/pathogens30100...
). Indeed, the Australian Dawn strain also showed large deletions in the omp8 gene (327 bp) and AM415 gene (1,194 bp). Pierlé et al. (2014)Pierlé S, Imaz-Rosshandler I, Kerudin A, Sambono J, Lew-Tabor A, Rolls P, et al. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens 2014; 3(1): 57-72. http://dx.doi.org/10.3390/pathogens3010057. PMid:25364572.
http://dx.doi.org/10.3390/pathogens30100...
suggested that the low virulence of the Dawn strain could be related to these genomic deletions. The observation of omp9 deletions in Brazilian strains suggests that omp9 size variability may be more common than previously assumed and that this does not constitute a geographical signature of the Australian strains. Moreover, the distinct size of the omp9 deletion among different strains might be associated with a highly polymorphic region.
Based on gene transcription level, it is plausible to hypothesize that OMP7, OMP8 and OMP9 proteins seem to be related to the stages of A. marginale in its vertebrate-host, mainly in persistently infected cattle (Noh et al., 2006Noh SM, Brayton KA, Knowles DP, Agnes JT, Dark MJ, Brown WC, et al. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect Immun 2006; 74(6): 3471-3479. http://dx.doi.org/10.1128/IAI.01843-05. PMid:16714578.
http://dx.doi.org/10.1128/IAI.01843-05...
). These genes are seen to have low transcription levels in tick cell cultures, in contrast to cultures performed in vertebrate cells. In addition, knockout of omp10, which also caused a reduction of omp9 expression, did not impair A. marginale viability in the ISE6 tick cell lineage. Indeed, it seems that membrane proteins of A. marginale from erythrocytes form a large complex constituted by OMP7, OMP8 and OMP9, arranged with MSP2, MSP3, MSP4, OMP1, OPAG2, AM779, AM780, AM1011, AM854 and VirB1 (Noh et al., 2008Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, et al. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect Immun 2008; 76(5): 2219-2226. http://dx.doi.org/10.1128/IAI.00008-08. PMid:18316389.
http://dx.doi.org/10.1128/IAI.00008-08...
). Meanwhile, the surface protein complex of A. marginale from tick cells was formed only by MSP2, MSP3, MSP4, AM778 and AM854. It is important to state that most of these results were obtained using in vitro model, and may not represent exactly what happens in the naturally infected organisms.
In this regard, this complex of surface proteins seems to play a key role in the antigenic/immunological response against A. marginale. Noh et al. (2013)Noh SM, Turse JE, Brown WC, Norimine J, Palmer GH. Linkage between Anaplasma marginale outer membrane proteins enhances immunogenicity but is not required for protection from challenge. Clin Vaccine Immunol 2013; 20(5): 651-656. http://dx.doi.org/10.1128/CVI.00600-12. PMid:23446216.
http://dx.doi.org/10.1128/CVI.00600-12...
showed that the antibody response induced by the linked surface complex was much greater than that induced by a pool of the proteins in non-assembled form. Since the host response to membrane antigens is strictly dependent on the composition of the surface complex, it is reasonable to hypothesize that the lack/deletion of one molecule could affect the immunological response of other surface proteins. Thus, as omp9 has been considered a vaccine candidate, and it is highly recognized by immune serum of cattle immunized with outer membrane fraction; it is possible to hypothesize that changes in gene sequence (or deletions) may affect the interaction of A. marginale and host immune system (Deringer et al., 2017Deringer JR, Forero-Becerra EG, Ueti MW, Turse JE, Futse JE, Noh SM, et al. Identification of a T-Cell Epitope That Is Globally Conserved among Outer Membrane Proteins (OMPs) OMP7, OMP8, and OMP9 of Anaplasma marginale Strains and with OMP7 from the A. marginale subsp. centrale Vaccine Strain. Clin Vaccine Immunol 2017; 24(1): e00406-e00416. http://dx.doi.org/10.1128/CVI.00406-16. PMid:27795302.
http://dx.doi.org/10.1128/CVI.00406-16...
). Hence, further studies involving omp9 genotyping in other A. marginale strains seem to be essential for improving the epidemiological data and elucidating the roles of this gene in infection.
The most studied genes/proteins of A. marginale are membrane proteins, particularly MSPs. They belong to a particular group of outer membrane proteins widely studied because their importance as immunodominant antigens. The proteins coded by the msp1 and msp2 superfamilies represent a significant proportion of the molecules expected on the surface of the organism. The MSP1, MSP2 and MSP3 proteins are immunodominant molecules to which most of the host immune response is targeted (Alleman et al., 1997Alleman AR, Palmer GH, McGuire TC, McElwain TF, Perryman LE, Barbet AF. Anaplasma marginale major surface protein 3 is encoded by a polymorphic, multigene family. Infect Immun 1997; 65(1): 156-163. http://dx.doi.org/10.1128/IAI.65.1.156-163.1997. PMid:8975906.
http://dx.doi.org/10.1128/IAI.65.1.156-1...
; Barbet et al., 1987Barbet AF, Palmer GH, Myler PJ, McGuire TC. Characterization of an Immunoprotective Protein Complex of Anaplasma marginale by Cloning and Expression of the Gene Coding for Polypeptide Am1O5L. Infect Immun 1987; 55(10): 2428-2435. http://dx.doi.org/10.1128/IAI.55.10.2428-2435.1987. PMid:2443451.
http://dx.doi.org/10.1128/IAI.55.10.2428...
; Blouin et al., 2003Blouin EF, Saliki JT, De la Fuente J, Garcia-Garcia JC, Kocan KM. Antibodies to Anaplasma marginale major surface proteins 1a and 1b inhibit infectivity for cultured tick cells. Vet Parasitol 2003; 111(2-3): 247-260. http://dx.doi.org/10.1016/S0304-4017(02)00378-3. PMid:12531299.
http://dx.doi.org/10.1016/S0304-4017(02)...
; Brown et al., 1998Brown WC, Shkap V, Zhu D, McGuire TC, Tuo W, McElwain TF, et al. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect Immun 1998; 66(11): 5406-5413. http://dx.doi.org/10.1128/IAI.66.11.5406-5413.1998. PMid:9784551.
http://dx.doi.org/10.1128/IAI.66.11.5406...
, 2001aBrown WC, McGuire TC, Zhu D, Lewin HA, Sosnow J, Palmer GH. Highly Conserved Regions of the Immunodominant Major Surface Protein 2 of the Genogroup II Ehrlichial Pathogen Anaplasma marginale Are Rich in Naturally Derived CD4+ T Lymphocyte Epitopes that Elicit Strong Recall Responses. J Immunol 2001a; 166(2): 1114-1124. http://dx.doi.org/10.4049/jimmunol.166.2.1114. PMid:11145692.
http://dx.doi.org/10.4049/jimmunol.166.2...
,bBrown WC, Palmer GH, Lewin HA, McGuire TC. CD4+ T lymphocytes from calves immunized with Anaplasma marginale major surface protein 1 (MSP1), a heteromeric complex of MSP1a and MSP1b, preferentially recognize the MSP1a carboxyl terminus that is conserved among strains. Infect Immun 2001b; 69(11): 6853-6862. http://dx.doi.org/10.1128/IAI.69.11.6853-6862.2001. PMid:11598059.
http://dx.doi.org/10.1128/IAI.69.11.6853...
, 2003Brown WC, Brayton KA, Styer CM, Palmer GH. The Hypervariable Region of Anaplasma marginale Major Surface Protein 2 (MSP2) Contains Multiple Immunodominant CD4+ T Lymphocyte Epitopes That Elicit Variant-Specific Proliferative and INF-γResponses in MSP2 Vaccinates. J Immunol 2003; 170(7): 3790-3798. http://dx.doi.org/10.4049/jimmunol.170.7.3790. PMid:12646645.
http://dx.doi.org/10.4049/jimmunol.170.7...
; Kocan et al., 2001Kocan KM, Halbur T, Blouin EF, Onet V, de la Fuente J, Garcia-Garcia JC, et al. Immunization of cattle with Anaplasma marginale derived from tick cell culture. Vet Parasitol 2001; 102(1-2): 151-161. http://dx.doi.org/10.1016/S0304-4017(01)00519-2. PMid:11705661.
http://dx.doi.org/10.1016/S0304-4017(01)...
; Oberle et al., 1988Oberle SM, Palmer GH, Barbet AF, McGuire TC. Molecular size variations in an immunoprotective protein complex among isolates of Anaplasma marginale. Infect Immun 1988; 56(6): 1567-1573. http://dx.doi.org/10.1128/IAI.56.6.1567-1573.1988. PMid:3372021.
http://dx.doi.org/10.1128/IAI.56.6.1567-...
; Palmer et al., 1994Palmer GH, Eid G, Barbet AF, McGuire TC, McElwain TF. The Immunoprotective Anaplasma marginale Major Surface Protein 2 Is Encoded by a Polymorphic Multigene Family. Infect Immun 1994; 62(9): 3808-3816. http://dx.doi.org/10.1128/IAI.62.9.3808-3816.1994. PMid:8063397.
http://dx.doi.org/10.1128/IAI.62.9.3808-...
).
Anaplasmosis persistent infection is a phenomenon in which A. marginale remains viable for long-term in cattle even after its clinical recovery. Carrier cattle maintain low-level of A. marginale, and may serve as a reservoir for vector infection. Also, this asymptomatic form, eventually, could be reverted to a clinical presentation depending on the host immune status (Kieser et al., 1990Kieser ST, Eriks IS, Palmer GH. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect Immun 1990; 58(4): 1117-1119. http://dx.doi.org/10.1128/IAI.58.4.1117-1119.1990. PMid:2318532.
http://dx.doi.org/10.1128/IAI.58.4.1117-...
). It is plausible that antigenic variation on major immunodominant proteins helps the parasite in this process of persistent survival in an immunocompetent host. In this sense, antigenic variation of MSP2, MSP3 and other MSP proteins may be part of this process. Brayton et al. (2003)Brayton KA, Meeus PFM, Barbet AF, Palmer GH. Simultaneous variation of the Immunodominant Outer Membrane Proteins, MSP2 and MSP3, during Anaplasma marginale persistence in vivo. Infect Immun 2003; 71(11): 6627-6632. http://dx.doi.org/10.1128/IAI.71.11.6627-6632.2003. PMid:14573687.
http://dx.doi.org/10.1128/IAI.71.11.6627...
demonstrated that simultaneous switching of MSP2 and MSP3 variants occurs during infection. Moreover, the ability of these two molecules to work in concert may serve to amplify the antigenic diversity of the surface coat, thus enabling the microorganism to survive the host immune response (Brayton et al., 2005Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 2005; 102(3): 844-849. http://dx.doi.org/10.1073/pnas.0406656102. PMid:15618402.
http://dx.doi.org/10.1073/pnas.040665610...
). A summary of the 147 SNPs found in the membrane protein genes, with comparison between the Brazilian strains, is shown in Table 2. Details of the INDELs identified in genes from surface proteins can be found in Table 3.
Single nucleotide polymorphisms (SNPs) in the surface protein genes from the Palmeira strain, compared with the Jaboticabal strain of A. marginale.
Insertions and deletions (INDELs) in the surface protein genes of the Palmeira strain, compared with the Jaboticabal strain of A. marginale.
Single nucleotide polymorphisms were found in the outer membrane protein genes: omp14 (39 SNPs), omp13 (25 SNPs), omp8 (17 SNPs) and omp1 and omp10 (10 SNPs each). They were also found in the major surface protein genes: msp1bpg2 (23 SNPs) and msp4 (one SNP). The MSP2, MSP3 and MSP4 proteins reside in the outer membrane with surface-exposed domains (Vidotto et al., 1994Vidotto MC, McGuire TC, McElwain TF, Palmer GH, Knowles DP Jr. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect Immun 1994; 62(7): 2940-2946. http://dx.doi.org/10.1128/IAI.62.7.2940-2946.1994. PMid:8005681.
http://dx.doi.org/10.1128/IAI.62.7.2940-...
). MSP2 and MSP3 are immunodominant proteins that are antigenically variable and serve to allow evasion of the host immune response (Brayton et al., 2003Brayton KA, Meeus PFM, Barbet AF, Palmer GH. Simultaneous variation of the Immunodominant Outer Membrane Proteins, MSP2 and MSP3, during Anaplasma marginale persistence in vivo. Infect Immun 2003; 71(11): 6627-6632. http://dx.doi.org/10.1128/IAI.71.11.6627-6632.2003. PMid:14573687.
http://dx.doi.org/10.1128/IAI.71.11.6627...
; Brown et al., 2003Brown WC, Brayton KA, Styer CM, Palmer GH. The Hypervariable Region of Anaplasma marginale Major Surface Protein 2 (MSP2) Contains Multiple Immunodominant CD4+ T Lymphocyte Epitopes That Elicit Variant-Specific Proliferative and INF-γResponses in MSP2 Vaccinates. J Immunol 2003; 170(7): 3790-3798. http://dx.doi.org/10.4049/jimmunol.170.7.3790. PMid:12646645.
http://dx.doi.org/10.4049/jimmunol.170.7...
; French et al., 1998French DM, McElwain TF, McGuire TC, Palmer GH. Expression of Anaplasma marginale Major Surface Protein 2 Variants during Persistent Cyclic Rickettsemia. Infect Immun 1998; 66(3): 1200-1207. http://dx.doi.org/10.1128/IAI.66.3.1200-1207.1998. PMid:9488414.
http://dx.doi.org/10.1128/IAI.66.3.1200-...
, 1999French DM, Brown WC, Palmer GH. Emergence of Anaplasma marginale antigenicvariants during persistent rickettsemia. Infect Immun 1999; 67(11): 5834-5840. http://dx.doi.org/10.1128/IAI.67.11.5834-5840.1999. PMid:10531237.
http://dx.doi.org/10.1128/IAI.67.11.5834...
; Meeus et al., 2003Meeus PFM, Brayton KA, Palmer GH, Barbet AF. Conservation of a gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol Microbiol 2003; 47(3): 633-643. http://dx.doi.org/10.1046/j.1365-2958.2003.03331.x. PMid:12535066.
http://dx.doi.org/10.1046/j.1365-2958.20...
).
The msp2 superfamily includes the members of the msp2, msp3 and msp4 gene families. The genome contains one full-length expression site for these genes. In addition, there are seven msp2 and seven msp3 functional pseudogenes in the St. Maries strain (Brayton et al., 2005Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 2005; 102(3): 844-849. http://dx.doi.org/10.1073/pnas.0406656102. PMid:15618402.
http://dx.doi.org/10.1073/pnas.040665610...
). The MSP1 protein is a surface-exposed heteromeric complex consisting of MSP1a and MSP1b. msp1α is a single-copy gene and exhibits differences among strains caused by variable numbers and sequences of tandem repeat units of 86-89 bp in length (Palmer et al., 2001Palmer GH, Rurangirwa FR, McElwain TF. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J Clin Microbiol 2001; 39(2): 631-635. http://dx.doi.org/10.1128/JCM.39.2.631-635.2001. PMid:11158120.
http://dx.doi.org/10.1128/JCM.39.2.631-6...
). MSP1b is encoded by a small multigene family of five genes, consisting of two full-length and three partial versions (pg) (Brayton et al., 2005Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 2005; 102(3): 844-849. http://dx.doi.org/10.1073/pnas.0406656102. PMid:15618402.
http://dx.doi.org/10.1073/pnas.040665610...
). The genes encoding MSP1a-like proteins 2 and 3 (mlp2 and mlp3) showed four and five SNPs, respectively. Six SNPs were found in the msp2 operon-associated gene 3 (opag3), three SNPs in the membrane protein terC and four in the outer membrane protein tolC (Table 2). Regarding the omp8 gene, there was a single nucleotide insertion causing a frameshift in the Palmeira strain (Table 3).
Polymorphisms found among strains from different continents can be related to different selection pressures, such as differences in management, treatment, prevention, host genetic constitution and arthropod vectors. While in Brazil the vectors are presumed to be primarily insects of the order Diptera and, also possibly, males of the cattle tick Rhipicephalus microplus (Estrada-Peña et al., 2006Estrada-Peña A, Bouattour A, Camicas J-L, Guglielmone A, Horak I, Jongejan F, et al. The known distribution and ecological preferences of the tick subgenus Boophilus (Acari : Ixodidae) in Africa and Latin America. Exp Appl Acarol 2006; 38(2-3): 219-235. http://dx.doi.org/10.1007/s10493-006-0003-5. PMid:16596355.
http://dx.doi.org/10.1007/s10493-006-000...
); in Australia the vector species is presumed to be Rhipicephalus australis (Burger et al., 2014Burger TD, Shao R, Barker SC. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol Phylogenet Evol 2014; 76: 241-253. http://dx.doi.org/10.1016/j.ympev.2014.03.017. PMid:24685498.
http://dx.doi.org/10.1016/j.ympev.2014.0...
); and in the United States the major vectors are the ticks Dermacentor andersoni, Dermacentor variabilis, and Dermacentor albipictus(Kocan et al., 2008Kocan KM, de la Fuente J, Blouin EF. Advances toward understanding the molecular biology of the Anaplasma-tick interface. Front Biosci 2008; 13(13): 7032-7045. http://dx.doi.org/10.2741/3208. PMid:18508714.
http://dx.doi.org/10.2741/3208...
). Importantly, previous studies have shown that the Jaboticabal strain is not transmitted transovarially in R. microplus ticks (Esteves et al., 2015Esteves E, Pohl PC, Klafke GM, Reck J, Fogaça AC, Martins JR, et al. Low temperature affects cattle tick reproduction but does not lead to transovarial transmission of Anaplasma marginale. Vet Parasitol 2015; 214(3-4): 322-326. http://dx.doi.org/10.1016/j.vetpar.2015.07.010. PMid:26255094.
http://dx.doi.org/10.1016/j.vetpar.2015....
). Moreover, widespread use of the live attenuated vaccine for A. centrale has been reported in Australia (Herndon et al., 2013Herndon DR, Ueti MW, Reif KE, Noh SM, Brayton KA, Agnes JT, et al. Identification of multilocus genetic heterogeneity in Anaplasma marginale subsp. centrale and its restriction following tick-borne transmission. Infect Immun 2013; 81(5): 1852-1858. http://dx.doi.org/10.1128/IAI.00199-13. PMid:23509140.
http://dx.doi.org/10.1128/IAI.00199-13...
) and southern Brazil, but not in the United States or in southeastern Brazil.
Another important influence may come from anaplasmosis treatment. Only two drugs are used in the United States, i.e, chlortetracycline and oxytetracycline (FDA, 2013US Food and Drug Administration – FDA. FDA approved animal drug products [online]. 2013 [cited 2021 Jan 4]. Available from: http://www.accessdata.fda.gov/scripts/animaldrugsatfda/
http://www.accessdata.fda.gov/scripts/an...
), while in Australia the commonly used drugs are imidocarb and oxytetracycline (Rogers & Dunster, 1984Rogers RJ, Dunster PJ. The elimination of Anaplasma marginale from carrier cattle by treatment with long acting oxytetracycline. Aust Vet J 1984; 61(9): 306. http://dx.doi.org/10.1111/j.1751-0813.1984.tb06026.x. PMid:6517790.
http://dx.doi.org/10.1111/j.1751-0813.19...
; Wilson et al., 1979Wilson AJ, Parker R, Parker M, Hall WT, Trueman KF. Chemotherapy of acute bovine anaplasmosis. Aust Vet J 1979; 55(2): 71-73. http://dx.doi.org/10.1111/j.1751-0813.1979.tb15167.x. PMid:444164.
http://dx.doi.org/10.1111/j.1751-0813.19...
). In Brazil, oxytetracycline, imidocarb and enrofloxacin are widely used for anaplasmosis treatment (Facury-Filho et al., 2012Facury-Filho EJ, de Carvalho AÚ, Ferreira PM, Moura MF, Apolinário BC, Santos LP, et al. Effectiveness of enrofloxacin for the treatment of experimentally-induced bovine anaplasmosis. Rev Bras Parasitol Vet 2012; 21(1): 32-36. http://dx.doi.org/10.1590/S1984-29612012000100007. PMid:22534942.
http://dx.doi.org/10.1590/S1984-29612012...
; Gonçalves, 2000Gonçalves PM. Epidemiologia e controle da Tristeza Parasitária Bovina na região sudeste do Brasil. Cienc Rural 2000; 30(1): 187-194. http://dx.doi.org/10.1590/S0103-84782000000100030.
http://dx.doi.org/10.1590/S0103-84782000...
; Gotze et al., 2008Gotze MM, Nizoli LQ, Silva SS. Efeitos da oxitetraciclina na recomposição do hematócrito de vacas leiteiras durante surto de anaplasmose bovina. Rev Bras Parasitol Vet 2008; 17(Suppl 1): 92-95. PMid:20059824.).
This paper presents the first genomes of the South American strains of A. marginale. These results suggest, considering the genomic information available at this moment, that Brazilian strains constitute one particular clade that is grouped in a larger American group together with North American strains, and is more distantly related to the Australian strains. These data contribute to better understanding of the evolutionary relationships of A. marginale strains. In addition, they show that there is a need for further studies on the genetic variability of other A. marginale strains, both in South America and in other regions, to obtain fuller epidemiological and pan-genome understanding.
Acknowledgements
The authors would like to thank Biomatters Ltd for granting a free license for the Geneious software; Dr. Eduardo Eizirik (FABIO, PUCRS, Porto Alegre, RS, Brazil); and members of the Arthropod Genomics and Evolution Laboratory (IB, USP, São Paulo, Brazil) for help and advice in bioinformatics analyses. Special thanks to Dr. João Ricardo Martins (IPVDF), Dr. Guilherme Klafke (IPVDF) and Dr. Sirlei Daffre (USP). This work was financially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Projeto SANIMARS (Financiadora de Estudos e Projetos, FINEP), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and INCT Entomologia Molecular.
-
How to cite: Dall’Agnol B, Webster A, Souza UA, Barbieri A, Mayer FQ, Cardoso GA, et al. Genomic analysis on Brazilian strains of Anaplasma marginale. Braz J Vet Parasitol 2021; 30(2): e000421. https://doi.org/10.1590/S1984-29612021043
References
- Alleman AR, Palmer GH, McGuire TC, McElwain TF, Perryman LE, Barbet AF. Anaplasma marginale major surface protein 3 is encoded by a polymorphic, multigene family. Infect Immun 1997; 65(1): 156-163. http://dx.doi.org/10.1128/IAI.65.1.156-163.1997 PMid:8975906.
» http://dx.doi.org/10.1128/IAI.65.1.156-163.1997 - Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998; 396(6707): 133-140. http://dx.doi.org/10.1038/24094 PMid:9823893.
» http://dx.doi.org/10.1038/24094 - Baêta BA, Ribeiro CCDU, Teixeira RC, Cabezas-Cruz A, Passos LMF, Zweygarth E, et al. Characterization of two strains of Anaplasma marginale isolated from cattle in Rio de Janeiro, Brazil, after propagation in tick cell culture. Ticks Tick Borne Dis 2015; 6(2): 141-145. http://dx.doi.org/10.1016/j.ttbdis.2014.11.003 PMid:25468764.
» http://dx.doi.org/10.1016/j.ttbdis.2014.11.003 - Barbet AF, Palmer GH, Myler PJ, McGuire TC. Characterization of an Immunoprotective Protein Complex of Anaplasma marginale by Cloning and Expression of the Gene Coding for Polypeptide Am1O5L. Infect Immun 1987; 55(10): 2428-2435. http://dx.doi.org/10.1128/IAI.55.10.2428-2435.1987 PMid:2443451.
» http://dx.doi.org/10.1128/IAI.55.10.2428-2435.1987 - Blouin EF, Saliki JT, De la Fuente J, Garcia-Garcia JC, Kocan KM. Antibodies to Anaplasma marginale major surface proteins 1a and 1b inhibit infectivity for cultured tick cells. Vet Parasitol 2003; 111(2-3): 247-260. http://dx.doi.org/10.1016/S0304-4017(02)00378-3 PMid:12531299.
» http://dx.doi.org/10.1016/S0304-4017(02)00378-3 - Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA 2005; 102(3): 844-849. http://dx.doi.org/10.1073/pnas.0406656102 PMid:15618402.
» http://dx.doi.org/10.1073/pnas.0406656102 - Brayton KA, Meeus PFM, Barbet AF, Palmer GH. Simultaneous variation of the Immunodominant Outer Membrane Proteins, MSP2 and MSP3, during Anaplasma marginale persistence in vivo. Infect Immun 2003; 71(11): 6627-6632. http://dx.doi.org/10.1128/IAI.71.11.6627-6632.2003 PMid:14573687.
» http://dx.doi.org/10.1128/IAI.71.11.6627-6632.2003 - Brown WC, Brayton KA, Styer CM, Palmer GH. The Hypervariable Region of Anaplasma marginale Major Surface Protein 2 (MSP2) Contains Multiple Immunodominant CD4+ T Lymphocyte Epitopes That Elicit Variant-Specific Proliferative and INF-γResponses in MSP2 Vaccinates. J Immunol 2003; 170(7): 3790-3798. http://dx.doi.org/10.4049/jimmunol.170.7.3790 PMid:12646645.
» http://dx.doi.org/10.4049/jimmunol.170.7.3790 - Brown WC, McGuire TC, Zhu D, Lewin HA, Sosnow J, Palmer GH. Highly Conserved Regions of the Immunodominant Major Surface Protein 2 of the Genogroup II Ehrlichial Pathogen Anaplasma marginale Are Rich in Naturally Derived CD4+ T Lymphocyte Epitopes that Elicit Strong Recall Responses. J Immunol 2001a; 166(2): 1114-1124. http://dx.doi.org/10.4049/jimmunol.166.2.1114 PMid:11145692.
» http://dx.doi.org/10.4049/jimmunol.166.2.1114 - Brown WC, Palmer GH, Lewin HA, McGuire TC. CD4+ T lymphocytes from calves immunized with Anaplasma marginale major surface protein 1 (MSP1), a heteromeric complex of MSP1a and MSP1b, preferentially recognize the MSP1a carboxyl terminus that is conserved among strains. Infect Immun 2001b; 69(11): 6853-6862. http://dx.doi.org/10.1128/IAI.69.11.6853-6862.2001 PMid:11598059.
» http://dx.doi.org/10.1128/IAI.69.11.6853-6862.2001 - Brown WC, Shkap V, Zhu D, McGuire TC, Tuo W, McElwain TF, et al. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect Immun 1998; 66(11): 5406-5413. http://dx.doi.org/10.1128/IAI.66.11.5406-5413.1998 PMid:9784551.
» http://dx.doi.org/10.1128/IAI.66.11.5406-5413.1998 - Burger TD, Shao R, Barker SC. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol Phylogenet Evol 2014; 76: 241-253. http://dx.doi.org/10.1016/j.ympev.2014.03.017 PMid:24685498.
» http://dx.doi.org/10.1016/j.ympev.2014.03.017 - da Silva JB, André MR, Machado RZ. Low genetic diversity of Anaplasma marginale in calves in an endemic area for bovine anaplasmosis in the state of São Paulo, Brazil. Ticks Tick Borne Dis 2016; 7(1): 20-25. http://dx.doi.org/10.1016/j.ttbdis.2015.07.018 PMid:26318264.
» http://dx.doi.org/10.1016/j.ttbdis.2015.07.018 - Dark MJ, Herndon DR, Kappmeyer LS, Gonzales MP, Nordeen E, Palmer GH, et al. Conservation in the face of diversity: multistrain analysis of an intracellular bacterium. BMC Genomics 2009; 10(1): 16. http://dx.doi.org/10.1186/1471-2164-10-16 PMid:19134224.
» http://dx.doi.org/10.1186/1471-2164-10-16 - de La Fuente J, Passos LMF, Van Den Bussche RA, Ribeiro MFB, Facury-Filho EJ, Kocan KM. Genetic diversity and molecular phylogeny of Anaplasma marginale isolates from Minas Gerais, Brazil. Vet Parasitol 2004; 121(3-4): 307-316. http://dx.doi.org/10.1016/j.vetpar.2004.02.021 PMid:15135871.
» http://dx.doi.org/10.1016/j.vetpar.2004.02.021 - de la Fuente J, Van Den Bussche RA, Garcia-Garcia JC, Rodríguez SD, García MA, Guglielmone AA, et al. Phylogeography of New World isolates of Anaplasma marginale based on major surface protein sequences. Vet Microbiol 2002; 88(3): 275-285. http://dx.doi.org/10.1016/S0378-1135(02)00122-0 PMid:12151201.
» http://dx.doi.org/10.1016/S0378-1135(02)00122-0 - de la Fuente J, Van Den Bussche RA, Prado TM, Kocan KM. Anaplasma marginale msp1α genotypes evolved under positive selection pressure but are not markers for geographic isolates. J Clin Microbiol 2003; 41(4): 1609-1616. http://dx.doi.org/10.1128/JCM.41.4.1609-1616.2003 PMid:12682152.
» http://dx.doi.org/10.1128/JCM.41.4.1609-1616.2003 - Deringer JR, Forero-Becerra EG, Ueti MW, Turse JE, Futse JE, Noh SM, et al. Identification of a T-Cell Epitope That Is Globally Conserved among Outer Membrane Proteins (OMPs) OMP7, OMP8, and OMP9 of Anaplasma marginale Strains and with OMP7 from the A. marginale subsp. centrale Vaccine Strain. Clin Vaccine Immunol 2017; 24(1): e00406-e00416. http://dx.doi.org/10.1128/CVI.00406-16 PMid:27795302.
» http://dx.doi.org/10.1128/CVI.00406-16 - Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol 2001; 51(Pt6): 2145-2165. http://dx.doi.org/10.1099/00207713-51-6-2145 PMid:11760958.
» http://dx.doi.org/10.1099/00207713-51-6-2145 - Esteves E, Pohl PC, Klafke GM, Reck J, Fogaça AC, Martins JR, et al. Low temperature affects cattle tick reproduction but does not lead to transovarial transmission of Anaplasma marginale. Vet Parasitol 2015; 214(3-4): 322-326. http://dx.doi.org/10.1016/j.vetpar.2015.07.010 PMid:26255094.
» http://dx.doi.org/10.1016/j.vetpar.2015.07.010 - Estrada-Peña A, Bouattour A, Camicas J-L, Guglielmone A, Horak I, Jongejan F, et al. The known distribution and ecological preferences of the tick subgenus Boophilus (Acari : Ixodidae) in Africa and Latin America. Exp Appl Acarol 2006; 38(2-3): 219-235. http://dx.doi.org/10.1007/s10493-006-0003-5 PMid:16596355.
» http://dx.doi.org/10.1007/s10493-006-0003-5 - Facury-Filho EJ, de Carvalho AÚ, Ferreira PM, Moura MF, Apolinário BC, Santos LP, et al. Effectiveness of enrofloxacin for the treatment of experimentally-induced bovine anaplasmosis. Rev Bras Parasitol Vet 2012; 21(1): 32-36. http://dx.doi.org/10.1590/S1984-29612012000100007 PMid:22534942.
» http://dx.doi.org/10.1590/S1984-29612012000100007 - Fournier PE, Dubourg G, Raoult D. Clinical detection and characterization of bacterial pathogens in the genomics era. Genome Med 2014; 6(11): 114. http://dx.doi.org/10.1186/s13073-014-0114-2 PMid:25593594.
» http://dx.doi.org/10.1186/s13073-014-0114-2 - French DM, Brown WC, Palmer GH. Emergence of Anaplasma marginale antigenicvariants during persistent rickettsemia. Infect Immun 1999; 67(11): 5834-5840. http://dx.doi.org/10.1128/IAI.67.11.5834-5840.1999 PMid:10531237.
» http://dx.doi.org/10.1128/IAI.67.11.5834-5840.1999 - French DM, McElwain TF, McGuire TC, Palmer GH. Expression of Anaplasma marginale Major Surface Protein 2 Variants during Persistent Cyclic Rickettsemia. Infect Immun 1998; 66(3): 1200-1207. http://dx.doi.org/10.1128/IAI.66.3.1200-1207.1998 PMid:9488414.
» http://dx.doi.org/10.1128/IAI.66.3.1200-1207.1998 - Gonçalves PM. Epidemiologia e controle da Tristeza Parasitária Bovina na região sudeste do Brasil. Cienc Rural 2000; 30(1): 187-194. http://dx.doi.org/10.1590/S0103-84782000000100030
» http://dx.doi.org/10.1590/S0103-84782000000100030 - Gotze MM, Nizoli LQ, Silva SS. Efeitos da oxitetraciclina na recomposição do hematócrito de vacas leiteiras durante surto de anaplasmose bovina. Rev Bras Parasitol Vet 2008; 17(Suppl 1): 92-95. PMid:20059824.
- Gouy M, Guindon S, Gascuel O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol Biol Evol 2010; 27(2): 221-224. http://dx.doi.org/10.1093/molbev/msp259 PMid:19854763.
» http://dx.doi.org/10.1093/molbev/msp259 - Harris RS. Improved pairwise alignment of genomic DNA State College: Penn State University Press; 2007.
- Herndon DR, Ueti MW, Reif KE, Noh SM, Brayton KA, Agnes JT, et al. Identification of multilocus genetic heterogeneity in Anaplasma marginale subsp. centrale and its restriction following tick-borne transmission. Infect Immun 2013; 81(5): 1852-1858. http://dx.doi.org/10.1128/IAI.00199-13 PMid:23509140.
» http://dx.doi.org/10.1128/IAI.00199-13 - Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28(12): 1647-1649. http://dx.doi.org/10.1093/bioinformatics/bts199 PMid:22543367.
» http://dx.doi.org/10.1093/bioinformatics/bts199 - Kieser ST, Eriks IS, Palmer GH. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect Immun 1990; 58(4): 1117-1119. http://dx.doi.org/10.1128/IAI.58.4.1117-1119.1990 PMid:2318532.
» http://dx.doi.org/10.1128/IAI.58.4.1117-1119.1990 - Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol 2010; 167(2-4): 95-107. http://dx.doi.org/10.1016/j.vetpar.2009.09.012 PMid:19811876.
» http://dx.doi.org/10.1016/j.vetpar.2009.09.012 - Kocan KM, de la Fuente J, Blouin EF. Advances toward understanding the molecular biology of the Anaplasma-tick interface. Front Biosci 2008; 13(13): 7032-7045. http://dx.doi.org/10.2741/3208 PMid:18508714.
» http://dx.doi.org/10.2741/3208 - Kocan KM, de la Fuente J, Cabezas-Cruz A. The genus Anaplasma: new challenges after reclassification. Rev Sci Tech 2015; 34(2): 577-586. http://dx.doi.org/10.20506/rst.34.2.2381 PMid:26601458.
» http://dx.doi.org/10.20506/rst.34.2.2381 - Kocan KM, Halbur T, Blouin EF, Onet V, de la Fuente J, Garcia-Garcia JC, et al. Immunization of cattle with Anaplasma marginale derived from tick cell culture. Vet Parasitol 2001; 102(1-2): 151-161. http://dx.doi.org/10.1016/S0304-4017(01)00519-2 PMid:11705661.
» http://dx.doi.org/10.1016/S0304-4017(01)00519-2 - Machado RZ, Silva JB, André MR, Gonçalves LR, Matos CA, Obregón D. Outbreak of anaplasmosis associated with the presence of different Anaplasma marginale strains in dairy cattle in the states of São Paulo and Goiás, Brazil. Rev Bras Parasitol Vet 2015; 24(4): 438-446. http://dx.doi.org/10.1590/S1984-29612015078 PMid:26648009.
» http://dx.doi.org/10.1590/S1984-29612015078 - Meeus PFM, Brayton KA, Palmer GH, Barbet AF. Conservation of a gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol Microbiol 2003; 47(3): 633-643. http://dx.doi.org/10.1046/j.1365-2958.2003.03331.x PMid:12535066.
» http://dx.doi.org/10.1046/j.1365-2958.2003.03331.x - Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, et al. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect Immun 2008; 76(5): 2219-2226. http://dx.doi.org/10.1128/IAI.00008-08 PMid:18316389.
» http://dx.doi.org/10.1128/IAI.00008-08 - Noh SM, Brayton KA, Knowles DP, Agnes JT, Dark MJ, Brown WC, et al. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect Immun 2006; 74(6): 3471-3479. http://dx.doi.org/10.1128/IAI.01843-05 PMid:16714578.
» http://dx.doi.org/10.1128/IAI.01843-05 - Noh SM, Turse JE, Brown WC, Norimine J, Palmer GH. Linkage between Anaplasma marginale outer membrane proteins enhances immunogenicity but is not required for protection from challenge. Clin Vaccine Immunol 2013; 20(5): 651-656. http://dx.doi.org/10.1128/CVI.00600-12 PMid:23446216.
» http://dx.doi.org/10.1128/CVI.00600-12 - Oberle SM, Palmer GH, Barbet AF, McGuire TC. Molecular size variations in an immunoprotective protein complex among isolates of Anaplasma marginale. Infect Immun 1988; 56(6): 1567-1573. http://dx.doi.org/10.1128/IAI.56.6.1567-1573.1988 PMid:3372021.
» http://dx.doi.org/10.1128/IAI.56.6.1567-1573.1988 - Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 2001; 293(5537): 2093-2098. http://dx.doi.org/10.1126/science.1061471 PMid:11557893.
» http://dx.doi.org/10.1126/science.1061471 - Palmer GH, Eid G, Barbet AF, McGuire TC, McElwain TF. The Immunoprotective Anaplasma marginale Major Surface Protein 2 Is Encoded by a Polymorphic Multigene Family. Infect Immun 1994; 62(9): 3808-3816. http://dx.doi.org/10.1128/IAI.62.9.3808-3816.1994 PMid:8063397.
» http://dx.doi.org/10.1128/IAI.62.9.3808-3816.1994 - Palmer GH, Rurangirwa FR, McElwain TF. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J Clin Microbiol 2001; 39(2): 631-635. http://dx.doi.org/10.1128/JCM.39.2.631-635.2001 PMid:11158120.
» http://dx.doi.org/10.1128/JCM.39.2.631-635.2001 - Pierlé S, Imaz-Rosshandler I, Kerudin A, Sambono J, Lew-Tabor A, Rolls P, et al. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens 2014; 3(1): 57-72. http://dx.doi.org/10.3390/pathogens3010057 PMid:25364572.
» http://dx.doi.org/10.3390/pathogens3010057 - Pohl AE, Cabezas-Cruz A, Ribeiro MFB, Silveira JAG, Silaghi C, Pfister K, et al. Detection of genetic diversity of Anaplasma marginale isolates in Minas Gerais, Brazil. Rev Bras Parasitol Vet 2013; 22(1): 129-135. http://dx.doi.org/10.1590/S1984-29612013000100024 PMid:24252959.
» http://dx.doi.org/10.1590/S1984-29612013000100024 - Ripma LA, Simpson MG, Hasenstab-Lehman KE. Geneious! Simplified genome skimming methods for phylogenetic systematic studies: a case study in Oreocarya (Boraginaceae). Appl Plant Sci 2014; 2(12): 1400062. http://dx.doi.org/10.3732/apps.1400062 PMid:25506521.
» http://dx.doi.org/10.3732/apps.1400062 - Rogers RJ, Dunster PJ. The elimination of Anaplasma marginale from carrier cattle by treatment with long acting oxytetracycline. Aust Vet J 1984; 61(9): 306. http://dx.doi.org/10.1111/j.1751-0813.1984.tb06026.x PMid:6517790.
» http://dx.doi.org/10.1111/j.1751-0813.1984.tb06026.x - Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, et al. Human-mouse alignments with BLASTZ. Genome Res 2003; 13(1): 103-107. http://dx.doi.org/10.1101/gr.809403 PMid:12529312.
» http://dx.doi.org/10.1101/gr.809403 - Silva JB, Cabezas-Cruz A, Fonseca AH, Barbosa JD, de la Fuente J. Infection of water buffalo in Rio de Janeiro Brazil with Anaplasma marginale strains also reported in cattle. Vet Parasitol 2014a; 205(3-4): 730-734. http://dx.doi.org/10.1016/j.vetpar.2014.09.009 PMid:25260335.
» http://dx.doi.org/10.1016/j.vetpar.2014.09.009 - Silva JB, Fonseca AH, Barbosa JD, Cabezas-Cruz A, de la Fuente J. Low genetic diversity associated with low prevalence of Anaplasma marginale in water buffaloes in Marajó Island, Brazil. Ticks Tick Borne Dis 2014b; 5(6): 801-804. http://dx.doi.org/10.1016/j.ttbdis.2014.06.003 PMid:25108778.
» http://dx.doi.org/10.1016/j.ttbdis.2014.06.003 - Silva JB, Gonçalves LR, Varani AM, André MR, Machado RZ. Genetic diversity and molecular phylogeny of Anaplasma marginale studied longitudinally under natural transmission conditions in Rio de Janeiro, Brazil. Ticks Tick Borne Dis 2015; 6(4): 499-507. http://dx.doi.org/10.1016/j.ttbdis.2015.04.002 PMid:25985719.
» http://dx.doi.org/10.1016/j.ttbdis.2015.04.002 - US Food and Drug Administration – FDA. FDA approved animal drug products [online]. 2013 [cited 2021 Jan 4]. Available from: http://www.accessdata.fda.gov/scripts/animaldrugsatfda/
» http://www.accessdata.fda.gov/scripts/animaldrugsatfda/ - Vidotto MC, Kano SF, Gregori F, Headley SA, Vidotto O. Phylogenetic analysis of Anaplasma marginale strains from Paraná State, Brazil, using the msp1α and msp4 genes. J Vet Med B Infect Dis Vet Public Health 2006; 53(9): 404-411. http://dx.doi.org/10.1111/j.1439-0450.2006.00984.x PMid:17062116.
» http://dx.doi.org/10.1111/j.1439-0450.2006.00984.x - Vidotto MC, McGuire TC, McElwain TF, Palmer GH, Knowles DP Jr. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect Immun 1994; 62(7): 2940-2946. http://dx.doi.org/10.1128/IAI.62.7.2940-2946.1994 PMid:8005681.
» http://dx.doi.org/10.1128/IAI.62.7.2940-2946.1994 - Wilson AJ, Parker R, Parker M, Hall WT, Trueman KF. Chemotherapy of acute bovine anaplasmosis. Aust Vet J 1979; 55(2): 71-73. http://dx.doi.org/10.1111/j.1751-0813.1979.tb15167.x PMid:444164.
» http://dx.doi.org/10.1111/j.1751-0813.1979.tb15167.x - Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2004; 2(3): e69. http://dx.doi.org/10.1371/journal.pbio.0020069 PMid:15024419.
» http://dx.doi.org/10.1371/journal.pbio.0020069
Publication Dates
-
Publication in this collection
28 May 2021 -
Date of issue
2021
History
-
Received
11 Jan 2021 -
Accepted
15 Mar 2021