Abstract
The objective of the present study was to estimate the allelic and genotypic frequencies of the polymorphisms E2FB (AY138588.1: c.305C> T), located in the leptin gene (LEP), and TG5 (X05380.1:g.-422C>T), located in the thyroglobulin gene (TG), and evaluate the association of these polymorphisms in crossbred cattle of seven distinct genetic groups with the following traits: slaughter weight (SW), hot carcass weight (HCW), hot carcass yield (HCY), carcass fat thickness (CFT), ribeye area (REA), marbling (MARM) and shear force (SF). The animals were genotyped using the PCR-RFLP (Polymorphism Chain Reaction-Restriction Fragment Length Polymorphism) technique, using 201 products obtained from F1 Caracu × Nellore, Angus × Nellore and Valdostana × Nellore cows, mated to Canchim, Caracu and Red Angus bulls (only Caracu × Nellore cows were used with Red Angus bulls). The allelic and genotypic frequencies were compared using the Chi-squared test. Associations between the genotype of each polymorphism and the traits were analyzed using the General Linear Model (GLM) of statistical software SAS. The least squares means of genotypes of the polymorphisms were compared using Student's t test. The E2FB polymorphism in the LEP gene was associated with CFT, showing the potential for use in national programs for genetic improvement of beef cattle, through the inclusion of SNP in genotyping commercial tests. The TG5 polymorphism in the TG gene was not associated with any of the evaluated traits and was considered ineffective for selection of beef cattle in Brazilian herds.
fat thickness; marker-assisted selection; meat tenderness; shear force; SNP
BREEDING, GENETIC AND REPRODUCTION
Association of polymorphisms in the leptin and thyroglobulin genes with meat quality and carcass traits in beef cattle
Thiago Dutra de CarvalhoI; Fabiane SiqueiraII; Roberto Augusto de Almeida Torres JúniorII; Sérgio Raposo de MedeirosII; Gelson Luís Dias FeijóII; Maury Dorta de Souza JuniorI; Isabella Maiumi Zaidan BlechaII; Cleber Oliveira SoaresII
IUniversidade Federal de Mato Grosso do Sul, Campo Grande/MS, Brazil
IIEmbrapa Gado de Corte, Campo Grande/MS, Brazil
ABSTRACT
The objective of the present study was to estimate the allelic and genotypic frequencies of the polymorphisms E2FB (AY138588.1: c.305C> T), located in the leptin gene (LEP), and TG5 (X05380.1:g.-422C>T), located in the thyroglobulin gene (TG), and evaluate the association of these polymorphisms in crossbred cattle of seven distinct genetic groups with the following traits: slaughter weight (SW), hot carcass weight (HCW), hot carcass yield (HCY), carcass fat thickness (CFT), ribeye area (REA), marbling (MARM) and shear force (SF). The animals were genotyped using the PCR-RFLP (Polymorphism Chain Reaction-Restriction Fragment Length Polymorphism) technique, using 201 products obtained from F1 Caracu × Nellore, Angus × Nellore and Valdostana × Nellore cows, mated to Canchim, Caracu and Red Angus bulls (only Caracu × Nellore cows were used with Red Angus bulls). The allelic and genotypic frequencies were compared using the Chi-squared test. Associations between the genotype of each polymorphism and the traits were analyzed using the General Linear Model (GLM) of statistical software SAS. The least squares means of genotypes of the polymorphisms were compared using Student's t test. The E2FB polymorphism in the LEP gene was associated with CFT, showing the potential for use in national programs for genetic improvement of beef cattle, through the inclusion of SNP in genotyping commercial tests. The TG5 polymorphism in the TG gene was not associated with any of the evaluated traits and was considered ineffective for selection of beef cattle in Brazilian herds.
Key Words: fat thickness, marker-assisted selection, meat tenderness, shear force, SNP
Introduction
In beef cattle, the type and carcass traits determine the value of the animal for both the producer and the meat industry. Subcutaneous fat is necessary to ensure the quality of the carcass after post-mortem cooling (Felício, 1997). Marbling is also important because it represents the intramuscular fat that contributes to succulence, flavor and tenderness of the meat (Crouse et al., 1989).
The LEP gene encodes leptin, which is produced by adipocytes and implicated in feed intake regulation, energy balance, fat deposition, meat quality and reproduction efficiency (Houseknecht et al., 1998; Geary et al., 2003). Polymorphisms in this gene have been previously studied and associated with economic traits in beef cattle (Pomp et al., 1997; Buchanan et al., 2002; Nkrumah et al., 2004; Schenkel et al., 2005; Lusk, 2007; Anton et al., 2011; Woronuk et al., 2012).
The TG gene encodes the protein thyroglobulin, which is a precursor of triiodothyronine and tetraiodothyronine, hormones that act on lipid metabolism. It has been proposed that the TG gene possibly affects fat deposition. A polymorphism at the 5' untranslated region of the gene was associated with the marbling score (Barendse, 1999) and included in commercial genotyping panels (Casas et al., 2005; Van Eenennaam, 2007).
However, before this information can be used in national genetic improvement programs, validation studies in different breeds and cattle populations, produced in Brazilian environmental conditions, are necessary. Very few validation data are available in Brazil. Furthermore, in most cases, the genetic markers assessed only partially explain the phenotypic variance found, whereas the effects of these genotypes may vary among different populations. It is important to consider the existence of epistatic interactions, pleiotropy, genetic linkage or segregation of other genes that make up the genome and that can display different combinations of those in which the marker was described. In addition, the interaction of the genotype with the environment should also be considered.
Therefore, the objective of the present study was to estimate the allelic and genotypic frequencies of single nucleotide polymorphisms (SNP) in the LEP and TG genes, and to assess associations between genotype, meat quality and carcass traits.
Material and Methods
The present study was carried out on an experimental farm at Embrapa Beef Cattle, Campo Grande - Mato Grosso do Sul, Brazil. Crossbred animals, born between 2007 and 2008, obtained from F1 Caracu × Nellore (CN), Angus × Nellore (AN) and Valdostana × Nellore (VN) cows, with Canchim (CC), Caracu (CR) and Red Angus (RA) bulls (RA used only with CN cows), were used, consisting of seven different genetic groups.
Experimental animals were subjected to the production system of super-precocious steers (14 to 16 months). This production system promotes the development of homogeneous carcasses and retail cuts with uniform fat cover, allowing better cooling in different parts of the carcass and increasing meat production by reducing the slaughter age of animals.
For each of the yearly assessments, animals produced in each season were separated into contemporary groups according to the season of birth: cattle born before October 15th formed contemporary groups; cattle born after October 16th formed other groups.
The selection of bulls for the production of the experimental population was performed according to the recommendations of the respective breed and associations of cattle breeders, and was based on the commercial availability of semen collected in artificial insemination centers. At least 10 bulls of each breed with the smallest possible degree of kinship were chosen.
In the finishing phase, the use of animals produced by artificial insemination was prioritized over animals born late in the season, which were product of natural mating on pasture. In total, 252 animals were produced and 201 weaning cattle were confined in stalls and subjected to an individual assessment of consumption until they reached the slaughter endpoint. The animals were slaughtered at around 15 months of age, when fat thickness in the dorsal-lumbar region (between the 12th and the 13th ribs) reached at least 5 millimeters (mm). During the finishing period, the animals were weighed every 28 days to monitor weight gain. They were also subjected to ultrasound examinations to assess the stage of carcass fat deposition.
The following two diets were provided for the animals: (A) 40% sorghum silage as forage, 53.2% corn grain and 3.7% soybean meal; (B) 40% sorghum silage as forage, 19.6% corn grain, 3.1% soybean meal with addition of 15% cottonseed and 20% soybean hulls. Within each genetic group, the animals were randomly distributed to the two diets. The daily feed supply was adjusted to 5-10% more than the actual intake, to characterize ad libitum consumption.
The animals were fasted and weighed at slaughter (SW). After slaughter, carcass assessments were performed to obtain the hot carcass weight (HCW) and the hot carcass yield (HCY) was calculated using the HCW/SW ratio.
Measurements of carcass fat thickness (CFT), ribeye area (REA) and marbling score (MARM), assessed in the longissimus dorsi muscle, were performed between the 12th and 13th ribs of the right side of the carcass, inside the cold room, twenty-four hours after slaughter, following the methodology proposed by Muller (1980). The minimum time on finishing (DCONF) to acquire fat thickness of at least 5 mm was also analyzed.
Longissimus dorsi muscle samples were extracted to assess the meat tenderness by shear force (SF) using a Warner-Bratzler Meat Shear Slot Blade, a dull V-shaped blade and a TA XT Plus texturometer (Muller, 1980). From each animal, one steak (2.5 cm in thickness) was removed, and from this, three cylindrical samples (1.25 cm diameter), extracted following the course of muscle fibers, were used. Each cylindrical sample was sent to the Warner-Bratzler twice, and the force required to shear the muscle fibers was established as the mean of six measurements (two for each cylindrical samples).
Blood samples from 201 animals were collected for the genotypic assessment by venipuncture in 4.5 mL vacuum tubes containing K3-EDTA. Genomic DNA was extracted from leukocytes as described by Regitano & Coutinho (2001). The integrity and concentration of DNA samples was assessed by agarose gel electrophoresis (0.8%) and spectrophotometry in NanoDrop (Thermo Scientific ND 1000).
The animals were genotyped for two SNP using Polymerase Chain Reaction - Restriction Fragment Length Polymorphisms (PCR-RFLP). One SNP was located in the LEP gene, mapped on chromosome four of cattle, and the other SNP was located in the TG gene, mapped on chromosome 14.
The SNP in the LEP gene was located at position 305 in exon 2, presently identified as E2FB (AY138588.1:c.305C>T), previously described by Buchanan et al. (2002). This polymorphism is responsible for the substitution of cytosine (allele C) by thymine (allele T), causing an arginine to cysteine exchange in the protein sequence.
The gene TG encodes the protein thyroglobulin, which is a precursor of hormones that affect the metabolism of lipids. Polymorphism TG5 (X05380.1:g.-422C>T), identified in the 5' untranslated region of the TG gene, is characterized by the transition of a cytosine (allele C) to a thymine (allele T) in the protein sequence (Barendse, 1999; Barendse et al., 2004). This polymorphism was patented by Barendse (1999), and is used in programs of Marker-Assisted Selection (MAS) to increase marbling levels, thus improving the meat quality of beef cattle.
The alleles C and T of SNP E2FB were detected by the amplification of fragments of 94 base pairs (bp) in exon 2 followed by digestion with Kpn2I, as described by Buchanan et al. (2002). The alleles C and T of SNP TG5 were detected by the amplification of fragments of 545 bp in the 5' untranslated region, followed by digestion with MboI (Barendse, 1999).
The allele and genotype frequencies for both SNP were determined for each genetic group, and compared using the Chi-Squared test, with a significance level of 5%. Deviations of genotype frequencies from those expected with the Hardy-Weinberg equilibrium were also analyzed considering a 5% significance level.
For association analysis, phenotypic traits related to meat quality and carcass were submitted to analysis of variance by the General Linear Model (GLM) procedure of the program SAS (Statistical Analysis System, version 9.2), using a model based on the effects of genotypes of each polymorphism, year of birth (season), cattle lot (according to season of birth), gender, genetic group of sire, cow genetic group and diet, as follows:
Yijklmnopq = µ + Ak + Ll (Ak) + Sm + GGn + GGto + GGvp + Dq + gj + εi,
in which yijklmnopq = the value observed in a specific variable in the i-th individual born in the k-th year and the l-th lot within each year, with the m-th gender and the n-th genetic group formed by crossing the o-th genetic group of sire with the p-th cow genetic group, which received the q-th diet and which showed the j-th genotype for a given locus; µ = overall mean for a specific variable y; Ak = effect of year of birth of the animal, in which k ranged from 2007-2008; Ll (Ak) = effect of the lot of animals within each year, in which l ranged from 1-2; Sm = effect of gender, wherein m assumes values male or female; GGn = effect of genetic group of the i-th individual, wherein n assumes values CCAN, CCCN, CCVN, CRAN, CRCN, CRVN or RACN; GGto = effect of genetic group of sire, where in o assumes values CC, CR or RA; GGvp = effect of cow genetic group, wherein p assumes values AN, CN or VN; Dq = effect of diet, wherein q assumes values A or B; gj = effect of the genotype of the animal for a specific locus, wherein j assumes values AA, Aa or aa, according to the homozygosity or heterozygosity status of the individual; εi = random error associated with each observation i.
The least square means and the standard error of the mean of the assessed traits for the two polymorphisms were estimated using Student's t-test. When significant effects on polymorphisms were observed (P<0.05), the allelic substitution effect was estimated by substituting the effect of the genotype of the marker in the statistical model for covariates representing the amount of the least frequent allele (0, 1 and 2).
Results and Discussion
Carcass traits and meat quality, features of multigene regulation, are economically important to both the beef industry and beef consumers. The molecular biology applied to cattle genetic improvement enables the identification, mapping and analyses of the regions that control quantitative traits (Quantitative Trait Loci - QTL), as well as the use of molecular markers (MAS) to increase the frequency of favorable alleles in a specific population (Stone et al., 2005).
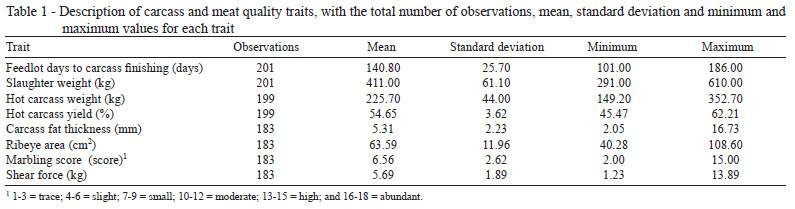
In order to improve the carcass traits and meat quality through genetic improvement of beef cattle, candidate genes such as the LEP and TG genes were studied and polymorphisms were mapped. In the present study, SNP previously described by Buchanan et al. (2002) and Barendse (1999) were assessed aiming to validate the markers in beef cattle produced under Brazilian conditions (Table 1).
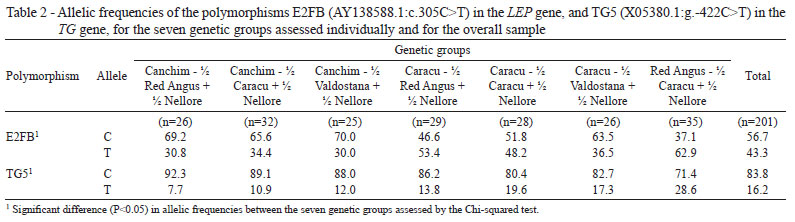
The alleles C and T of polymorphism E2FB were observed in all of the genetic groups studied. Restriction fragments of 19 and 75 bp were found in genotype CC. On the other hand, only fragments of 94 bp were detected in genotype TT. Heterozygous animals (CT) exhibited the three fragments (19, 75 and 94 bp). Using the Chi-squared test, significant differences (P<0.05) were found in allelic and genotypic frequencies among the seven genetic groups (Tables 2 and 3, respectively).
Frequencies of 56.7% (C) and 43.3% (T) were found in the seven genetic groups assessed (Table 2). Similar results have been previously reported for the same polymorphism (Buchanan et al., 2002). These authors analyzed a sample of 154 Bos taurus taurus (Angus, Charolais, Hereford and Simmental) and found allelic frequencies of 54% for allele C and 46% for allele T. Allele T was considered favorable to carcass fat deposition (Buchanan et al., 2002). From the 201 bovine samples analyzed in the present study, 59 exhibited the CC genotype, with 110 of genotype CT and 32 of genotype TT. The genetic group CCVN was the only group in which animals with the TT genotype were not found (Table 3).
Fortes et al. (2009) assessed the association of polymorphisms in the LEP (AY138588.1:c.305C>T), TG (X05380.1:g.-422C>T) and DGAT1 (AJ318490:c.[10433G>A; 10434C>A]) genes with fat thickness, total lipids, marbling, ribeye area and shear force in 147 animals from five distinct genetic groups (Nellore, Rubia Gallega × Nellore, Canchim, and three-way cross Brangus and Braunvieh). Frequencies of 81% (allele C) and 19% (allele T) were found for the polymorphism E2FB. The lower frequencies for the allele T were observed in cattle with a higher Bos taurus indicus genetic composition (4.3% Nellore and 7.7% Rubia Gallega × Nellore). In the genetic three-way cross groups (Brangus and Braunvieh), similar frequencies for allele T (26.3% and 20%, respectively) were detected. These frequencies were different from those found in the Canchim group (39%). No significant associations were found between the polymorphism E2FB and the phenotypic traits analyzed.
In the present study, genetic groups with a higher Bos taurus indicus composition (CCVN, CCAN and CCCN) also exhibited lower frequencies of allele T (30%, 30.8% and 34.4%, respectively) (Table 2). On the other hand, genetic groups with a higher Bos taurus taurus composition (CRVN, CRCN, CRAN, CRCN and RACN) exhibited frequencies of 36.5%, 48.2%, 53.5% and 62.9%, respectively. These results confirm the observations of Buchanan et al. (2002), who reported a frequency of 58% for allele T in SNP E2FB in Angus cattle. Nkrumah et al. (2004) reported a frequency of 71% for the same allele in 144 Angus crosses.
Schenkel et al. (2005) studied five different polymorphisms in the gene LEP (UASMS1, UASMS2, UASMS3, E2JW and E2FB) using 1,111 Bos taurus taurus crosses (Angus, Charolais, Limousin and Simmental) and found a frequency of 61% for allele T in SNP E2FB. No significant differences were found (Chi-squared test) beteween the allelic frequencies of any polymorphism analyzed in cattle with a genetic composition > 5/8 for a particular Bos taurus taurus breed. However, the animals with a higher Angus genetic composition exhibited a higher frequency for the allele T in SNP E2FB than the other groups. Thus, previously reported frequencies for allele T in SNP E2FB (Buchanan et al., 2002; Nkrumah et al., 2004; Schenkel et al., 2005), are very similar to those found in the present study for the genetic group RACN (62.9%).
With respect to the polymorphism TG5, both allelic variants (C and T) were found in all of the genetic groups assessed. Significant differences (Chi-squared test) in allelic and genotypic frequencies were found between the seven groups (P<0.05) (Tables 2 and 3, respectively).
Allele T of SNP TG5 is recognized for enabling marbling and carcass fat deposition (Barendse, 1999; Thaller et al., 2003; Barendse et al., 2004; Fortes et al., 2009; Pannier et al., 2010). Of the 201 animals studied, 142 exhibited the genotype CC, 53 the genotype CT and six the genotype TT. In the genetic groups CRAN and CRVN, the genotype TT was not detected (Table 3). In the global sample, allele C was found with a frequency of 83.8%, whereas the frequency of allele T was 16.2% (Table 2). Only the genetic group CCVN was not in Hardy-Weinberg equilibrium (Table 3), with a 5% significance level.
The groups presenting a higher Bos taurus indicus genetic composition (CCAN, CCCN and CCVN), showed the lowest frequencies for allele T (7.7%, 10.9% and 12%, respectively). The groups with a higher Bos taurus taurus genetic composition (CRAN, CRVN, CRCN and RACN) exhibited frequencies of 13.8%, 17.3%, 19.6% and 28.6%, respectively (Table 2). These results are in accordance with those found by Casas et al. (2005), who reported a low frequency of allele T in SNP TG5 in Brahman cattle, as well as with the observations of Fortes et al. (2009), who reported that allele T (SNP TG5) is fixed in the Nellore breed. These authors concluded that the increase in allele T frequency was associated with the increased Bos taurus taurus genetic composition in the population studied: Rubia Gallega × Nellore (1.9%), three-way cross Brangus (15.8%), Canchim (22%) and three-way cross Braunvieh (33.3%). Similar frequencies have been previously reported (22% to 25%) in Bos taurus taurus (Thaller et al., 2003; Moore et al., 2003).
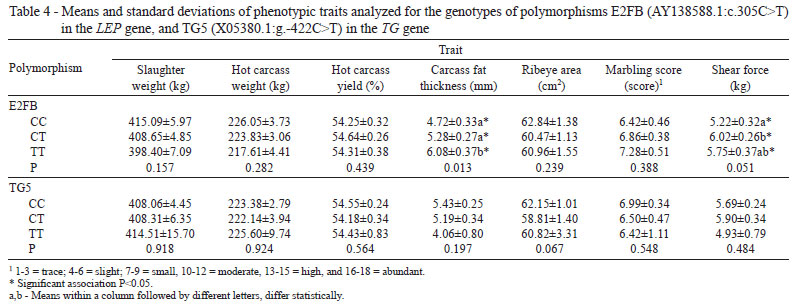
A significant association (P=0.013) was found between the polymorphism E2FB and fat thickness (Table 4), with an average effect of allele substitution of 0.6602±0.2238 mm (P=0.004) for allele T (Table 5). These results demonstrated that fat thickness increases according to the number of copies of allele T in the genotype (CT or TT). The difference between the genotypes TT and TC was greater than 0.80 cm2, whereas the difference between genotypes TT and CC was greater than 1.36 cm2, resulting in a superiority of the TT genotype over the others. These findings are in agreement with those described by Buchanan et al. (2002), who found a correlation between an increase in fat deposition and allele T in SNP E2FB.
For the shear force trait, the association with SNP E2FB was very close to the significance level (P=0.051) (Table 4). An association between shear force and SNP E2JW and E2FB was previously reported by Schenkel et al. (2005), analyzing five markers located in the leptin gene (UASMS1, UASMS2, UASMS3, E2JW and E2FB), as in the experiment described above.
Studies have demonstrated the presence of QTL for fat deposition and marbling in the chromosome 14 of cattle (Casas et al., 2000; Moore et al., 2003). Genes located in this QTL, as well as allelic variations in these genes, are important for efficient marker-assisted selection in commercial herds of beef cattle.
Although some studies have demonstrated the effect of the polymorphism TG5 on the marbling and fat thickness traits (Barendse, 1999; Casas et al., 2005; Shin & Chung, 2007; Anton et al., 2011), no statistically significant associations for slaughter weight (SW), hot carcass weight (HCW), hot carcass yield (HCY), carcass fat thickness (CFT), ribeye area (REA), marbling (MARM) and shear force (SF) were found in the present study. These results agree with those found by Moore et al. (2003), Rincker et al. (2006), Van Eenemmaam et al. (2007), Fortes et al. (2009) and Pannier et al. (2010).
According to Pannier et al. (2010), results related to associations between the TG gene polymorphisms and marbling and fat deposition traits are inconsistent. The authors estimated the allelic and genotype frequencies of TG5 polymorphisms in 459 cattle of various breeds (Limousin, Charolais, Friesian, Simmental, Angus, Hereford, Belgian Blue, Blonde d'Aquitaine and Salers) and investigated the association of this polymorphism in intramuscular fat deposition in the longissimus thoracis et lumborum and semimembranosus muscles in 138 crossbred cattle and found no significant associations.
Van Eenemmaam et al. (2007) performed validation studies of the commercial panel of molecular markers (GeneSTAR Quality Grade) for the marbling trait using 409 crossbred (Charolais × Angus) and 324 Hereford cattle and found no trait × genotype associations. This panel of molecular markers is composed of SNP TG5, QG2 and an anonymous SNP (unpublished data).
The results obtained in the present study suggest that other SNP in the TG gene, as well as other genes in the centromeric region of chromosome 14 should be studied in order to determine associations with the lipid metabolism in beef cattle. Validation studies of cattle produced in Brazilian environmental conditions are necessary to generate essential information for the incorporation of the DNA test to national genetic assessment programs.
Conclusions
Polymorphism E2FB, located in the leptin gene, was associated with fat thickness, showing the potential for use in national programs to improve beef cattle, by the inclusion of SNP in commercial genotyping panels. Polymorphism TG5, located in the thyroglobulin gene, was not associated with the phenotypic traits assessed, and was shown to be ineffective for the improvement of beef cattle herds in Brazil. However, these results do not exclude the possibility that other alleles of the TG gene may be associated with marbling or subcutaneous fat deposition, and do not exclude the centromeric region of chromosome 14 as a candidate region in the search for new genes and/or polymorphisms that can be used in genetic improvement programs for cattle.
Acknowledgments
The authors thank Sistema Embrapa de Gestão (Macroprograma 1 Projeto Bife de Qualidade 01.05.1.02.00.00 and Macroprograma 3 Projeto 03.08.01.020.00.00) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq- Processo Nº 506016/2008-0) for the financial support; and Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul (FUNDECT) for the Master's fellowship granted.
Received November 3, 2011 and accepted June 19, 2012
Corresponding author: fabiane@cnpgc.embrapa.br
- ANTON, I.; KOVÁCS, K.; HOLLÓ, G. et al. Effect of leptin, DGAT1 and TG gene polymorphisms on the intramuscular fat of Angus cattle in Hungary. Livestock Science, v.135, p.300-303, 2011.
- BARENDSE, W.J. Assessing lipid metabolism Patent. International Publication Number: WO 99/23248. World Intellectual Property Organization, Geneva, Switzerland, 1999.
- BARENDSE, W.J.; BUNCH, R.; THOMAS, M. et al. The TG5 thyroglobulin gene test for a marbling quantitative trait loci evaluated in feedlot cattle. Australian Journal of Experimental Agriculture, v.44, p.669-674, 2004.
- BUCHANAN, F.C.; FITZSIMMONS, C.J.; VAN KESSEL, A.G. et al. Association of a missense mutation in the bovine leptin gene with carcass fat content and leptin mRNA levels. Genetic Selection Evolution, v.34, p.105-116, 2002.
- CASAS, E.; WHITE, S.N.; RILEY, D.G. et al. Assessment of single nucleotide polymorphisms in genes residing on chromosomes 14 and 29 for association with carcass composition traits in Bos indicus cattle. Journal of Animal Science, v.83, p.13-19, 2005.
- CASAS, E.; SHACKELFORD, S.D.; KEELE, J.W. et al. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternative forms of myostatin. Journal of Animal Science, v.78, p.560-569, 2000.
- CROUSE, J.D.; CUNDIFF, L.V.; KOCH, R.M. et al. Comparisons of Bos indicus and Bos taurus inheritance for carcass beef characteristics and meat palatability. Journal of Animal Science, v.67, n.10, p.2661-2668, 1989.
- FELÍCIO, P.E. Fatores ante o post mortem que influenciam na qualidade da carne bovina. In: PEIXOTO, A.M.; MOURA, J.C.; FARIA, V.P. (Eds.) Produção do novilho de corte Piracicaba: Fundação de Estudos Agrários "Luis de Queiroz", 1997. p.79-97.
- FORTES, M.R.S.; CURI, R.A.; CHARDULO, L.A.L. et al. Bovine gene polymorphisms related to fat deposition and meat tenderness. Genetics and Molecular Biology, v.32, p.75-82, 2009.
- GEARY, T.W.; McFADIN, E.L.; MacNEIL, M.D. et al. Leptin as a predictor of carcass composition in beef cattle. Journal of Animal Science, v.81, p.1-8, 2003.
- HOUSEKNECHT, K.L.; BAILE, C.A.; MATTERI, R.L. et al. The biology of leptin: a review. Journal of Animal Science, v.76, p.1405-1420, 1998.
- LUSK, J.L. Association of single nucleotide polymorphism in the leptin gene with body weight and backfat growth curve parameters for beef cattle. Journal of Animal Science, v.85, p.1865-1872, 2007.
- MOORE, S.S.; LI, C.; BASARAB, J. et al. Fine mapping of quantitative trait loci and assessment of positional candidate genes for backfat on bovine chromosome 14 in a commercial line of Bos taurus Journal of Animal Science, v.81, p.1919-1925, 2003.
- MULLER, L. Normas para avaliação de carcaças e concurso de carcaças de novilhos Santa Maria: Universidade Federal de Santa Maria, 1980. v.1, 31p.
- NKRUMAH, J.D.; LI, C.; BASARAB, J.B. et al. Association of a single nucleotide polymorphism in the bovine leptin gene with feed intake, feed efficiency, growth, feeding behavior, carcass quality and body composition. Canadian Journal of Animal Science, v.84, p.211-219, 2004.
- PANNIER, L.; MULLEN, A.M.; HAMILL, R.M. et al. Association analysis of single nucleotide polymorphisms in DGAT1, TG and FABP4 genes and intramuscular fat in crossbred Bos taurus cattle. Meat Science, v.85, p.515-518, 2010.
- POMP, D.; ZOU, T.; CLUTTER, A.C. et al. Rapid communication: Mapping of leptin to bovine chromosome 4 by linkage analysis of a PCR-based polymorphism. Journal of Animal Science, v.75, p.1427, 1997.
- REGITANO, L.C.A.; COUTINHO, L.L. Biologia molecular aplicada à produção animal Brasília: Embrapa Informação Tecnológica, 2001. p.205.
- RINCKER, C.B.; PYATT, N.A.; BERGER, L.L. et al. Relationship among GeneSTAR marbling marker, intramuscular fat deposition, and expected progeny differences in early weaned Simmental steers. Journal of Animal Science, v.84, p.686-93, 2006.
- SCHENKEL, F.S.; MILLER, S.P.; YE, X. et al. Association of single nucleotide polymorphisms in the leptin gene with carcass and meat quality traits of beef cattle. Journal of Animal Science, v.83, p.2009-2020, 2005.
- SHIN, S.C.; CHUNG, E.R. Association of SNP marker in the Thyroglobulin gene with carcass and meat quality traits in Korean cattle. Asian-Australian Journal of Animal Science, v.20, p.172-177, 2007.
- STONE, R.T.; CASAS, E.; SMITH, T.P. et al. Identification of genetic markers for fat deposition and meat tenderness on bovine chromosome 5: development of a low-density single nucleotide polymorphism map. Journal of Animal Science, v.83, p.2280-2288, 2005.
- THALLER, G.; KÜHN, C.; WINTER, A. et al. DGAT1, a new positional and functional candidate gene for intramuscular fat deposition in cattle. Animal Genetics, v.34, p.354-357, 2003.
- VAN EENENNAAM, A.L.; LI, J.; THALLAMAN, R.M. et al. Validation of commercial DNA tests for quantitative beef quality traits. Journal of Animal Science, v.85, p.891-900, 2007.
- WORONUK, G.N.; MARQUESS, F.L.; JAMES, S.T. et al. Rapid communication: Association of leptin genotypes with beef cattle characteristics. Animal Genetics, v.43, p.608-610, 2012.
Publication Dates
-
Publication in this collection
20 Nov 2012 -
Date of issue
Oct 2012
History
-
Received
03 Nov 2011 -
Accepted
19 June 2012