Abstracts
The method used by YAGYU et al. for the subtype-specific polymerase chain reaction (PCR) amplification of the gp41 transmembrane region of the human immunodeficiency virus type-1 (HIV-1) env gene, was tested. HIV-1 proviral DNA from 100 infected individuals in Itajaí, South Brazil was used to analyze this method. Seventy individuals were determined according to this method as having PCR products at the expected size for subtypes B, C, D and F. Of these individuals, 26 (37.1%) were observed as having the expected amplification for subtype C, and 42 (60%) were observed as having the expected products for subtypes B and D. Of the subtype B and D amplicons, 16 (22.9%) were classified as subtype D, and 26 (37.1%) were classified as subtype B. Two individuals (2.9%) had amplicons that were observed after subtype F-specific amplification was performed. Sequencing and comparing the patient sequences to reference sequences confirmed the classification of sequences of subtypes C and B. However, sequences that were falsely determined as being D and F in the PCR assay were determined as being subtypes C and B, respectively, by sequence analysis. For those individuals from whom no amplified products were obtained, a low viral load that was indicated in their patient history may explain the difficulty in subtyping by PCR methods. This issue was demonstrated by the results of ANOVA when testing the effect of viral load on the success of PCR amplification. The alignment of the obtained sequences with HIV-1 reference sequences demonstrated that there is high intra-subtype diversity. This indicates that the subtype-specific primer binding sites were not conserved or representative of the subtypes that are observed in the Brazilian populations, and that they did not allow the correct classification of HIV-1 subtypes. Therefore, the proposed method by YAGYU et al. is not applicable for the classification of Brazilian HIV-1 subtypes.
HIV-1; gp41; Viral load; Subtypes; PCR; South Brazil
A metodologia para amplificação subtipo-específica por PCR da região transmembrana do gene env (gp41) do HIV-1, descrita por Yagyu e colaboradores, foi testada a partir de DNA proviral de 100 pacientes infectados pelo HIV-1 de Itajaí, Sul do Brasil. Setenta indivíduos apresentaram produtos amplificados e correspondentes aos subtipos B, C, D e F de acordo com a metodologia escolhida. Destes indivíduos, 26 (37,1%) apresentaram a amplificação esperada para o subtipo C de acordo com a metodologia; 42 (60%) apresentaram os produtos esperados para os subtipos B e D, sendo que na etapa seguinte de diferenciação destes subtipos, 16 (22,9%) corresponderam ao subtipo D e 26 (37,1%) ao subtipo B. Dois indivíduos (2,9%) mostraram produtos amplificados após a amplificação específica para o subtipo F. O sequenciamento e a comparação com sequências referências confirmou a subtipagem de HIV-1 C e B obtida pela metodologia. No entanto, indivíduos subtipados erroneamente como HIV-1 D e F pela metodologia, foram classificados pela comparação com sequências referências como subtipos C e B, respectivamente. Em relação aos indivíduos que não mostraram produtos amplificados, a baixa carga viral observada no histórico destes pacientes seria em parte responsável pela dificuldade na subtipagem pela metodologia de PCR, como demonstrado pelo resultado significativo no ANOVA ao testar o efeito da carga viral no sucesso da amplificação. O alinhamento das sequências obtidas com sequências referências de HIV-1 correspondentes à região da gp41 demonstrou que há uma alta diversidade intra-subtipo e que as regiões a partir das quais foram desenhados os oligonucleotídeos iniciadores HIV-1 subtipo-específicos não são conservadas nem suficientemente representativas dos subtipos observados nas populações brasileiras para permitir sua correta identificação. Portanto, esta metodologia não é aplicável para populações virais brasileiras.
INTRODUCTION
Human immunodeficiency viruses types 1 and 2 (HIV-1 and HIV-2), which are responsible for the Acquired Immunodeficiency Syndrome (AIDS), were introduced into the human population during the 20th century. In the Americas, the first recognized case of HIV-1 subtype B was observed in 19811010. Heeney JL, Dalgleish AG, Weiss RA. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313(5786):462-6.. From that first observed case until June 2011 more than six hundred thousand cases of AIDS have been detected in Brazil. Approximately thirty thousand of these cases were observed in the Santa Catarina State of South Brazil1515. Ministério da Sáude. Brasil. Boletim Epidemiológico AIDS e DST. Brasília; Ministério da Saúde; 2011.. A large number of reported cases are likely to be from the city of Itajaí in South Brazil. Port cities, such as Itajaí, are subject to a large influx of people from all around the world. Therefore, these cities are a main point of entry for new strains of HIV, resulting in the increased genetic diversity of HIV1313. Martinez AM, Barbosa EF, Ferreira PC, Cardoso FA, Silveira J, Sassi G, et al. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev Soc Bras Med Trop. 2002;35:471-6.,1414. Martinez AM, Hora VP, Santos AL, Mendoza-Sassi R, Von Groll A, Soares EA, et al. Determinants of HIV-1 mother-to-child transmission in Southern Brazil. An Acad Bras Cienc. 2006;78:113-21..
HIV-1 strains are divided into different groups, subtypes, and sub-subtypes according to their sequence. The main group (M) includes the majority of subtypes that are circulating in the human population (A, B, C, D, F, G, H, J and K), including approximately fifty circulating recombinant forms (CRFs) of HIV-1 that have been described worldwide1111. Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18:182-92.,1818. Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55-6.. The viral env gene encodes a precursor glycoprotein gp160 that is processed intracellularly to produce the native form of the envelope complex. The envelope complex comprises three gp120 surface subunits and three gp41 transmembrane subunits that are associated with each other by noncovalent interactions. The composition of the gp120-gp41 complex is related to the ability of the virus to infect host cells55. Esté JA, Telenti A. HIV entry inhibitors. Lancet. 2007;370(9581):81-8.,2121. Sanders RW, Korber B, Lu M, Berkhout B, Moore JP. Mutational analyses and natural variability of the gp41 ectodomain. 2002 HIV Sequence Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2002compendium.html
http://www.hiv.lanl.gov/content/sequence...
. The env gene is observed to have highly variable domains that are interspersed between conserved regions of gp120. These variable regions are distinctive of the HIV-1 subtypes44. Costa SM, Schechter M, Shindo N, Vicente AC, Oliveira EF, Pinto ME, et al. Sequence and phylogenetic analysis of glycoprotein 120 of an HIV type 1 variant (GWGR) prevalent in Brazil. AIDS Res Hum Retroviruses. 1995;11:1143-5.,1717. Pieniazek D, Chunfu Y, Lal RB. Phylogenetic analysis of gp41 envelope of HIV-1 groups M, N and O strains provides an alternate region for subtype determination. The Human retroviruses and Aids 1998 Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/98compendium.html
http://www.hiv.lanl.gov/content/sequence...
. The high level of heterogeneity observed in the gp120 coding region led to the analysis of the gp41 coding region as an alternative region for the identification of HIV-1 subtypes using short nucleotide sequences11. Agwale SM, Robbins KE, Odama L, Saekhou A, Zeh C, Edubio A, et al. Development of an env gp41-based heteroduplex mobility assay for rapid human immunodeficiency virus type 1 subtyping. J Clin Microbiol. 2001;39:2110-4.. Phylogenetic studies demonstrated that the gp41 sequence and the C2-V3 nucleotide sequence of gp120 showed concordance in the classification of HIV-1 subtypes11. Agwale SM, Robbins KE, Odama L, Saekhou A, Zeh C, Edubio A, et al. Development of an env gp41-based heteroduplex mobility assay for rapid human immunodeficiency virus type 1 subtyping. J Clin Microbiol. 2001;39:2110-4.,1717. Pieniazek D, Chunfu Y, Lal RB. Phylogenetic analysis of gp41 envelope of HIV-1 groups M, N and O strains provides an alternate region for subtype determination. The Human retroviruses and Aids 1998 Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/98compendium.html
http://www.hiv.lanl.gov/content/sequence...
. Additionally, conserved regions in the gp41 coding region were identified, allowing the design of primers for subtype-specific amplification and identification of HIV-12929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23..
HIV diversity is an important issue for disease progression and transmission, the diagnosis and measurement of viral load, response to antiretroviral therapy and drug resistance, as well as immune response and vaccine development1111. Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18:182-92.,1717. Pieniazek D, Chunfu Y, Lal RB. Phylogenetic analysis of gp41 envelope of HIV-1 groups M, N and O strains provides an alternate region for subtype determination. The Human retroviruses and Aids 1998 Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/98compendium.html
http://www.hiv.lanl.gov/content/sequence...
,2727. Tupinambás U, Ribeiro FA, Aleixo A, Greco D. Treatment switch guided by HIV-1 genotyping in Brazil. Braz J Infect Dis. 2006;10:82-8.. The diversity of HIV is also important for recognizing geographic boundaries of subtype distributions, identifying modes and routes of transmissions and identifying new recombinant forms of HIV1313. Martinez AM, Barbosa EF, Ferreira PC, Cardoso FA, Silveira J, Sassi G, et al. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev Soc Bras Med Trop. 2002;35:471-6.,1414. Martinez AM, Hora VP, Santos AL, Mendoza-Sassi R, Von Groll A, Soares EA, et al. Determinants of HIV-1 mother-to-child transmission in Southern Brazil. An Acad Bras Cienc. 2006;78:113-21..
YAGYU et al. 2828. Yagyu F, Ikeda Y, Ariyoshi K, Sugiura W, Wongkhomthong SA, Masuda M, et al. Differentiation of subtypes B and E of human immunodeficiency virus type 1 by polymerase chain reaction using novel env gene primers. J Virol Methods. 2002;101:11-20.,2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. described a rapid and simple method for the identification of HIV-1 subtypes by PCR amplification of a region of gp41 using subtype-specific primers. Therefore, our aim was to test whether the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. could classify HIV-1 subtypes in seropositive individuals from the population of Itajaí (Santa Catarina, South Brazil).
MATERIAL AND METHODS
1. Population and sampling: The Laboratório Municipal de Itajaí (LMI) is a laboratory of the Brazilian Network for CD4+/CD8+ Cell Count and HIV Viral Load, located in the city of Itajaí, Santa Catarina in South Brazil. The LMI is a reference laboratory for highly complex and diagnostic exams that serves patients from Itajaí and neighboring towns. However, HIV subtyping is only possible when genotyping is performed in the Lacen laboratory of Florianópolis, which is the main city of the Santa Catarina State. Patients located in Itajaí who were observed in the clinic for the routine examination of CD4+/CD8+ T cell counting and plasma HIV viral load were invited to participate in this research. Individuals who volunteered (n = 100) were over 18 years of age, gave their approval for the use of their samples and signed an informed consent. This research was approved by the Ethical Committee of the Universidade do Vale do Itajaí (UNIVALI).
Viral load (highest recorded, last recorded and mean of the last six exams) for each patient (n = 98) was determined by the NASBA (Nuclisens, Bioméreux) method. The viral load for each patient was used for a comparison with the HIV-1 subtype that was detected by PCR amplification using the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.. Patients also completed a form indicating their age and the mode by which they believed they were infected. Blood samples were collected at the LMI laboratory in tubes that contained EDTA between October and December of 2006.
2. DNA extraction: Blood samples were centrifuged at 2,500 × g for 15 min at room temperature. The plasma was removed and the buffy coat was washed with 2:1 ACK solution (0.15 M NH4Cl; 0.01 M KHCO3; 0.1 mM Na2EDTA) several times until all red blood cells were removed. After each wash, the cells were centrifuged at 500 × g for five min. The samples were then resuspended in PBS (8.09 mM Na2HPO4; 0.14 M NaCl; 1.47 mM KH2PO4; 2.68 mM KCl) and centrifuged at 500 × g for five min. The supernatant was then removed. These samples were transported on ice to the Molecular Genetics Laboratory of the University (UNIVALI), where they were stored at -75 °C until genomic DNA extraction was performed. DNA extraction was performed by resuspending the pellet in 700 µL of lysis buffer (10 mM Tris-HCl, pH 7.4; 5 mM EDTA, pH 8.0; 20% SDS). Subsequently, 25 µL of Proteinase-K (20 mg/mL) was added, and the mixture was incubated at 57 °C for 2-3 h. After the incubation was complete, Phenol/Chloroform purification was performed. The DNA was precipitated using chilled, absolute ethanol and resuspended in 30 µL of TE buffer (10 mM Tris-HCl, pH 7.4; 1 mM EDTA, pH 8.0).
3. Amplification by HIV-1 subtype-specific primers and sequencing: The gp41 region from the proviral HIV-1 DNA was amplified by PCR using subtype-specific primers and methods as described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. (Table 1). This method consists of sequential, nested PCR amplification steps that result in the amplification of different DNA fragments of specific sizes (Table 1) based on the HIV-1 subtype. For example, the BECO5 and BECO3 primers are the first round primers for all of the HIV-1 subtypes. For the nested PCR, the first PCR product is used as the template, the forward primer is the BE-ANCH primer, and the specificity of amplification is determined by the reverse primers B-SPEC, C-SPEC, E-SPEC or F-SPEC (Table 1). For the BE-ANCH:B-SPEC and BE-ANCH:E-SPEC primer pairs, a third amplification step must be performed to distinguish the B and D and the A, E and G subtypes, respectively (Table 1). The first PCR product is always used as the template after perceiving the expected fragment in the second-round PCR.
Primer sequences and amplification steps for HIV-1 subtype-specific amplification following the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.
Each 25 µL PCR reaction contained 1 × PCR buffer, 1.5 mM MgCl2, 1 µM of each primer, 0.2 mM of each dNTP and 1 U Taq DNA polymerase. The PCR template was composed of 1 µL of proviral DNA for the first PCR amplification, and 1 µL of the first-round PCR product was diluted 1:10 for all subsequent PCR amplification steps. The thermocycler was programmed as follows: 1 cycle of three min at 94 °C, followed by 35 cycles of one min at 94 °C, 1.5 min at 50 °C, and 1.5 min at 72 °C, and a final cycle of three min at 72 °C. The PCR products were perceived after horizontal electrophoresis using a 1.5% agarose gel and ethidium bromide staining. Molecular markers of 50 bp and 100 bp were used as reference markers.
DNA fragments obtained sizes that were close to the expected length after subtype-specific amplification from two individuals of each subtype and a representative for all of the subtype-specific fragments that were amplified were selected for sequencing. These fragments were isolated from the gel and purified using Ultrafree-DA (Millipore) filter units by centrifuging the column containing the DNA for 13 min at 5,000 × g. DNA was precipitated by ethanol precipitation and resuspension in water. The DNA was then sent to the sequencing laboratory of Macrogen, Inc. company, Korea (www.macrogen.com) for sequence analysis. Sequencing analysis was conducted under BigDye™ terminator cycling conditions. The reacted products were purified using ethanol precipitaion and were run using Automatic Sequencer 3730XL.
4. Data analysis: The sequences were edited using the CHROMAS v.2.31 (Technelysium Pty Ltd, 2005) software. The alignment of the sequences to the subtype reference sequences that were obtained from the online HIV database (http://hiv-web.lanl.gov) was performed using the ClustalW online tool (http://bioweb.pasteur.fr/seqanal/interfaces/clustalw-simple.html). Similarities between the patient sequences that were obtained in this study to other sequences in the NCBI HIV database (National Center for Biotechnology Information) were determined using the BLAST online tool (http://ncbi.nlm.nih.gov//BLAST). To verify the subtypes that were classified, a maximum likelihood phylogenetic tree was constructed based on the Tamura-Nei model using env reference sequences from the HIV database to determine the phylogenetic locations of the sequences from the patients that were subtyped using the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.. Confidence of the branches was evaluated by bootstrapping 500 replications. These analyses were conducted using Mega version 52525. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol Biol Evol. 2011;28:2731-9.. HIV-1 g41 proviral sequences were named according to HIV-1 nomenclature guidelines1818. Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55-6. and submitted to GenBank.
The effect of the viral load for each patient on the amplification of the gp41 region by the PCR method as described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. was evaluated by an ANOVA test after normalizing the viral load data by a log transformation. The statistical package STATISTICA for Windows, version 5.1 (Statsoft, Inc.), was used to determine the mean viral load and standard error. This software was also used to log-transform these data and for the ANOVA test of significance (p < 0.05).
RESULTS
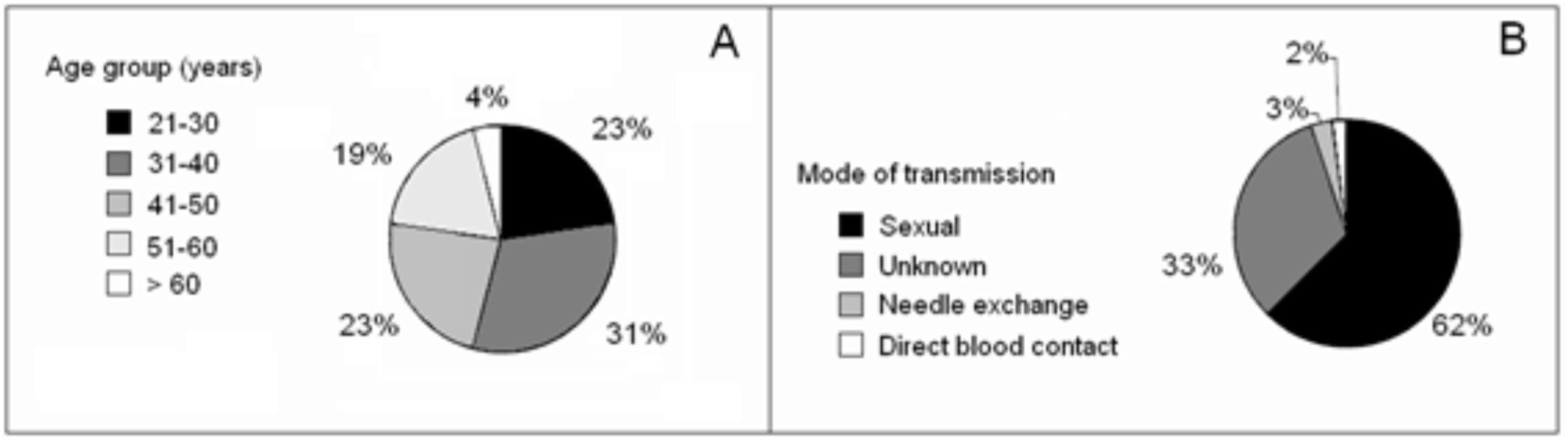
1. Population: The age and mode of HIV infection indicated by each patient are presented in Fig. 1. The average age of the participants was 40 ± 9.6 years; 53% of the patients were female and 47% were male (Fig. 1A). The predominant mode of transmission indicated by the patients was sexual transmission. A large number of patients (33%) did not know the mode by which they were infected (Fig. 1 B).
2. Viral load of patients and detection of HIV subtypes by PCR: In general, higher viral loads were observed in male patients than in female patients (Table 2). However, the viral load of the males had a high level of variability, and the differences in viral load between males and females were not significant (Last viral load, p = 0.331; mean viral load, p = 0.136; highest viral load, p = 0.061).
The successful amplification of subtype-specific products by PCR was reduced for patients with a lower viral load. This held true when the viral load was measured as the last recorded viral load of the patient, as the mean of the last six viral loads obtained, or as the highest viral load recorded for the history of the patient. Overall, the individuals whose proviral sequences could not be PCR-amplified were those patients who had lower viral loads (Fig. 2, Table 2). When comparing patients whose proviral sequences could be PCR-amplified with patients whose proviral sequences could not be amplified, an ANOVA significance test confirmed that there was a significant difference between the mean value of the viral load of the last six exams or for the highest value of the viral load recorded between these two patient populations (Table 2, Fig. 2). In accordance with these data, it was not possible to subtype individuals whose highest viral load was less than 60,000 copies/mL or who had a mean value that was less than 30,000 copies/mL.
Detection of HIV-1 subtype by PCR amplification compared to viral load measurements for each patient. The viral load was calculated as the last recorded viral load obtained from the patient (lower panel), as the mean of the last six viral load measurements (middle panel) and as the highest value (upper panel) that was recorded in the patient history.
3. Identification of subtype by the PCR testing method: The analysis of the PCR products that were obtained after the subtype-specific amplification indicated the presence of numerous non-specific amplicons. Nonetheless, the annealing temperature was maintained at 50 °C as described previously. PCR products at the expected size for the different HIV-1 subtypes were obtained for seventy individuals using the previously described method2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.. None of the amplified sequences corresponded to subtypes A, E or G (Table 3). For 26 (37.1%) patients, an amplified fragment of 676 bp or 698 bp was observed, which is close to the expected size (697 bp) for subtype C-specific amplification. For 42 (60%) individuals, an amplified fragment of 442 bp was observed, which is close to the expected size for the subtype B/D strains. Among these 42 patients, the expected 350-bp fragment after D-subtype specific amplification was observed for 16 (22.9%) patients. Therefore, the remaining 26 individuals (37.1%) who were not observed to have specific amplification using subtype D-specific primers were classified as subtype B. An amplified fragment of 782 bp was observed for two individuals, which was close to the expected size (778 bp) after subtype F-specific amplification (Table 3).
Summary of the results of HIV-1 subtyping following subtype-specific PCR determination and analysis of DNA fragments that were close in size to the expected fragment for different subtypes as described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.. The viral sequences obtained in this study were compared to HIV-1 reference strains and entered into GenBank with the correct HIV-1 subtypes
4. Comparing patient sequences to the HIV-1 subtype reference sequences: All of the sequences that were classified as subtypes B and C were correctly assigned to their respective HIV-1 subtypes after being aligned with the HIV-1 reference sequences and clustered in their respective subtype branches of the phylogenetic tree that was constructed (Fig. 3). Therefore, the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. was able to correctly identify sequences that were subtypes B and C (Table 3, Fig. 3). However, the sequences that were classified as subtypes D and F according to the method by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. were incorrectly classified. The sequences that were classified as subtype D clustered phylogenetically with the subtype C reference sequences with 100% confidence (bootstrap value). Both of the subtype F sequences that were classified by the YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. method were more similar to the subtype B reference sequences and clustered with the subtype B branch of the phylogenetic tree (Fig. 3).
Maximum likelihood phylogenetic analysis of env reference sequences with Itajaí city patient sequences (in bold, see Table 2) that were classified by the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.. HIV-1 subtypes that were predicted by this method are indicated in brackets. Reference sequences of the env gene subtypes that were obtained from the HIV-1 database are represented by the corresponding subtype and the GenBank accession number. Numbers on the branches represent the confidence of the branches that were obtained by 500 bootstrap replications.
The subtype C proviral amplicons that were correctly assigned by the method of YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. were of two different sizes when amplified by the subtype-specific primers. One amplicon, identified as 06BR.IJ003, was of 698 bp, very close in size to the expected fragment. The second 676-bp fragment was 21 bp shorter and was identified as 06BR.IJ019 (Table 3). When looking for similarities in NCBI database, it was discovered that the 676-bp fragment (06BR.IJ019) shared a 93% sequence identity with a recombinant BC subtype (GenBank accession number EF091932) and a 92% sequence identity with two subtype C viruses (GenBank accession number AY563170 and AB254148). This fragment clustered with a 99% confidence interval within the subtype C and recombinant subtype BC branches of the phylogenetic tree of the HIV reference sequences (Fig. 3). It also grouped with the subtype C sequence (82% confidence) that corresponded to a Brazilian male from the city of Porto Alegre in South Brazil that was recorded in 199277. Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651-67.. The other subtype C sequence (06BR. IJ003) had a 93% sequence identity with other subtype C viruses (GenBank accession numbers AY727526, U09126 and U15121) and also showed a 93% identity to a subtype A virus (GenBank accession number AY751057). In the phylogenetic analysis (Fig. 3), the 06BR.IJ003 sequence was clustered within the subtype C branch with a 99% degree of confidence.
A total of 98-99% of the subtype D sequence (06BR.IJ039) showed a 92% sequence identity with subtype C viruses (GenBank accession number U39234 and AY727527) and with a virus that did not have a specified subtype (GenBank accession number U39238). By phylogenetic analysis, this sequence clustered together with a subtype C sequence (06BR.IJ003) with 99% confidence (Fig. 3). The second subtype D sequence (06BR.IJ093) also showed high sequence identity (92-93%) with subtype C viruses (GenBank accession numbers AY669740, AY727525 and U39234) and with a subtype BC recombinant virus (GenBank accession number EF091932). This sequence was clustered with 100% confidence with the branch in the phylogenetic tree that contained all of the subtype C reference sequences that were included in the analysis (Fig. 3).
Both of the subtype F sequences that were classified (06BR. IJ026 and 06BR.IJ083) were 4 bp longer (782 pb) than the expected fragment size and were more similar to subtype B and recombinant subtype BF reference sequences. A total of 97-100% of the fragment sequences showed a 91% sequence identity with subtype B (GenBank accession number AY247225) and recombinant subtype BF (GenBank accession number DQ358811) viruses. In the phylogenetic tree of HIV reference sequences, both sequences clustered with each other with 100% confidence but clustered on the subtype B branch with only 36% confidence (Fig. 3).
5. Degree of specificity of the subtype-specific primers described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.: The analysis of the sequence alignments against subtype references from the HIV database (http://hiv-web.lanl.gov) showed that the subtype-specific primers that were described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. were not 100% specific for all of the variants for the respective subtypes that were registered in the database (Fig. 4). For this figure, dots represent sites where there is an agreement between the primer sequences described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. and sequences in the database, and the bases in black indicate sites where there were differences in the sequences. For example, under stringent amplification conditions, the forward BEANCH primer would not have effectively recognized two of the subtype B, two of the subtype D, one subtype F1, and all four of the subtype C reference strains that were included in this analysis (Fig. 4). The same is true for the reverse primers indicated in Fig. 4, which also have divergent sequences in comparison with some of the subtype reference sequences. It was also observed that some of the subtype-specific primers amplified other HIV-1 subtypes using this method2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.. This was specifically observed in the amplification of variants of subtypes C and B using the subtype D- and F-specific primers, respectively.
Alignment of the primer binding sites for the primers described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. which were used for HIV subtype-specific amplification of gp41 for this study. For the alignment, fragments that were obtained in this study are compared to reference sequences for subtypes B, C, D and F1 that were obtained from the HIV-1 database. Dots represent sequence homology with the original primer sequence described by YAGYU et al. The bases in black within the primer region represent divergent sequences from the respective primer sequence region. Underlined sequences represent the superpositions of primers.
DISCUSSION
As a consequence of their rapid in vivo turnover rate and frequent recombination events, the high mutation rates that are observed for HIV have resulted in high levels of genetic variability1616. Peeters M. Recombinant HIV sequences: their role in the global epidemic. 2000 HIV Sequence Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2000compendium.html
http://www.hiv.lanl.gov/content/sequence...
,2222. Santos AF, Schrago CG, Martinez AM, Mendoza-Sassi R, Silveira J, Sousa TM, et al. Epidemiologic and evolutionary trends of HIV-1 CRF31_BC-related strains in southern Brazil. J Acquir Immune Defic Syndr. 2007;45:328-33. and an increasing variety of HIV-1 subtypes. The identification of predominant subtypes, the distribution of subtypes in the country, and the identification of new HIV-1 variants in the Brazilian population have been intensified to study the use of more efficient therapeutic drug treatments and for the development of HIV-1 vaccines. Therefore, researchers from developing countries are persistently looking for new, low-cost and efficient methods for subtype HIV-1. A promising method was described by YAGYU et al.
2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. that is predicated on the PCR amplification of the gp41 subunit of the env gene using subtype-specific primers. While this method showed satisfactory results in samples from HIV-1-infected individuals in the populations of Japan and Thailand, it did not show the same success using samples from the population of South Brazil.
This method failed to correctly assign certain variants of C and B subtypes that were identified in the Brazilian populations. The primers that were described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. were not specific for some subtype variants of HIV-1 that were observed in the Brazilian populations because the primer binding regions were not adequately conserved to facilitate the subtype-specific amplification. The lack of subtype-specific primer binding explains the need to maintain less stringent amplification conditions (annealing temperature of 50 °C) using this method. However, the use of non-stringent amplification conditions led to the amplification of non-specific subtypes. Conversely, increasing the annealing temperature will result in a failure to amplify many of the subtype variants. As shown in Fig. 4, the primer binding regions are neither conserved nor representative enough to be used for the identification of Brazilian HIV-1 subtypes. This is a major detriment for using this method for subtype HIV-1 infected individuals from regions that maintain high levels of HIV-1 genetic diversity. High intra-subtype variation in the gp41 subunit was also observed. This raises the question of whether gp41 contains conserved regions that could allow different subtypes to be recognized.
Thirty percent of individuals could not be subtyped by the PCR method that was used in this study because of a failure to amplify subtype-specific PCR products. While a detriment to the specificity of the PCR amplification, the less stringent conditions used in this method should facilitate the amplification of all patient samples. Another subtype-specific PCR method has also failed to amplify patient viral sequences (20%)1212. Locateli D, Stoco PH, Queiroz AT, Alcântara LC, Ferreira LG, Zanetti CR, et al. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in Southern Brazil. J Med Virol. 2007;79:1455-63.; therefore, this issue may be related to a history of low viral load for the patient (determined by plasma RNA levels) rather than to the PCR amplification method. This argument is supported by data that showed a significant effect of the viral load of the patient on the success of the subtype-specific amplification in this study (Fig. 2, Table 2). The highest viral load recorded for the patient and the mean viral load of the patient's last six exams were evaluated in this study. Therefore, mean lifetime viral loads that are less than 30,000 copies/mL for a patient will significantly reduce the possibility of identifying the HIV-1 subtype that has infected an individual.
Studies have demonstrated that the frequency of various subtypes is dependent on geographical location. HIV-1 molecular epidemiology studies have already detected four subtypes in the Brazilian population: B, C, D, and F99. Guimarães ML, dos Santos Moreira A, Loureiro R, Galvão-Castro B, Morgado MG. High frequency of recombinant genomes in HIV type 1 samples from Brazilian southeastern and southern regions. AIDS Res Hum Retroviruses. 2002;18:1261-9.,1313. Martinez AM, Barbosa EF, Ferreira PC, Cardoso FA, Silveira J, Sassi G, et al. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev Soc Bras Med Trop. 2002;35:471-6.,2323. Santos AF, Soares MA. HIV genetic diversity and drug resistance. Viruses. 2010;2:503-31.. The HIV-1 B subtype is the most prevalent and has shown a decreasing frequency since 1993 (94% in São Paulo) to the period 1999-2001 (64% in Rio de Janeiro)2020. Sabino EC, Diaz RS, Brigido LF, Learn GH, Mullins JI, Reingold AL, et al. Distribution of HIV-1 subtypes seen in an AIDS clinic in Sao Paulo City, Brazil. AIDS. 1996;10:1579-84.,2626. Teixeira SL, Bastos FI, Telles PR, Hacker MA, Brigido LF, de F Oliveira CA, et al. HIV-1 infection among injection and ex-injection drug users from Rio de Janeiro, Brazil: prevalence, estimated incidence and genetic diversity. J Clin Virol. 2004;31:221-6. to 2004 (23% in Santa Catarina)1212. Locateli D, Stoco PH, Queiroz AT, Alcântara LC, Ferreira LG, Zanetti CR, et al. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in Southern Brazil. J Med Virol. 2007;79:1455-63.. The frequency of subtype F viruses in the Brazilian population is highly dependent on the geographical location. Subtype F has been observed at a very low frequency in Northeast Brazil (2.7%)66. Gadelha SR, Shindo N, Cruz JN, Morgado MG, Galvão-Castro B. Molecular epidemiology of human immunodeficiency virus-1 in the state of Ceará, Northeast, Brazil. Mem Inst Oswaldo Cruz. 2003;98:461-3., in the extreme of South Brazil (3%)1313. Martinez AM, Barbosa EF, Ferreira PC, Cardoso FA, Silveira J, Sassi G, et al. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev Soc Bras Med Trop. 2002;35:471-6. and in São Paulo (6%)2020. Sabino EC, Diaz RS, Brigido LF, Learn GH, Mullins JI, Reingold AL, et al. Distribution of HIV-1 subtypes seen in an AIDS clinic in Sao Paulo City, Brazil. AIDS. 1996;10:1579-84.. However, subtype F has been observed at moderate frequencies in Rio de Janeiro both in the period 1994-1997 (24.4%)88. Guimarães ML, Bastos FI, Telles PR, Galvão-Castro B, Diaz RS, Bongertz V, et al. Retrovirus infections in a sample of injecting drug users in Rio de Janeiro City, Brazil: prevalence of HIV-1 subtypes, and co-infection with HTLV-I/II. J Clin Virol. 2001;21:143-51. and in the period 1999-2001 (16.7%)2626. Teixeira SL, Bastos FI, Telles PR, Hacker MA, Brigido LF, de F Oliveira CA, et al. HIV-1 infection among injection and ex-injection drug users from Rio de Janeiro, Brazil: prevalence, estimated incidence and genetic diversity. J Clin Virol. 2004;31:221-6.. Individuals that were infected with subtype C virus were identified at a frequency of 22% in 1997 in South Brazil1313. Martinez AM, Barbosa EF, Ferreira PC, Cardoso FA, Silveira J, Sassi G, et al. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev Soc Bras Med Trop. 2002;35:471-6.. In 2004 in the Santa Catarina State, subtype C virus was identified at a frequency of 48%1212. Locateli D, Stoco PH, Queiroz AT, Alcântara LC, Ferreira LG, Zanetti CR, et al. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in Southern Brazil. J Med Virol. 2007;79:1455-63.. In South Brazil, it was discovered in 2006 that 70% of HIV positive pregnant women were infected with C virus1414. Martinez AM, Hora VP, Santos AL, Mendoza-Sassi R, Von Groll A, Soares EA, et al. Determinants of HIV-1 mother-to-child transmission in Southern Brazil. An Acad Bras Cienc. 2006;78:113-21. and that subtype C was at a frequency of 50% of the HIV-1-infected subjects in South Brazil1414. Martinez AM, Hora VP, Santos AL, Mendoza-Sassi R, Von Groll A, Soares EA, et al. Determinants of HIV-1 mother-to-child transmission in Southern Brazil. An Acad Bras Cienc. 2006;78:113-21.,2323. Santos AF, Soares MA. HIV genetic diversity and drug resistance. Viruses. 2010;2:503-31.. Further studies showed that some HIV-positive individuals who were previously reported to be infected with subtype C virus were actually infected with the recombinant CRF31_BC variant2222. Santos AF, Schrago CG, Martinez AM, Mendoza-Sassi R, Silveira J, Sousa TM, et al. Epidemiologic and evolutionary trends of HIV-1 CRF31_BC-related strains in southern Brazil. J Acquir Immune Defic Syndr. 2007;45:328-33.,2424. Santos AF, Sousa TM, Soares EA, Sanabani S, Martinez AM, Sprinz E, et al. Characterization of a new circulating recombinant form comprising HIV-1 subtypes C and B in southern Brazil. AIDS. 2006;20:2011-9., representing 21.2% of the previously detected subtype C infection2222. Santos AF, Schrago CG, Martinez AM, Mendoza-Sassi R, Silveira J, Sousa TM, et al. Epidemiologic and evolutionary trends of HIV-1 CRF31_BC-related strains in southern Brazil. J Acquir Immune Defic Syndr. 2007;45:328-33.. Both subtypes A and D are extremely rare, and they are detected only after several years of analysis33. Couto-Fernandez JC, Eyer-Silva WA, Guimarães ML, Chequer-Fernandez SL, Grinsztejn B, Delaporte E, et al. Phylogenetic analysis of Brazilian HIV type 1 subtype D strains: tracing the origin of this subtype in Brazil. AIDS Res Hum Retroviruses. 2006;22:207-11..
Therefore, the 40% frequency of the subtype B virus (37.1% recognized correctly by the method by YAGYU et al. in addition to the 2.9% that was correctly identified as subtype B by sequencing) is higher than the frequencies that were identified for the city of Florianopolis in Santa Catarina State (23%) in 20041212. Locateli D, Stoco PH, Queiroz AT, Alcântara LC, Ferreira LG, Zanetti CR, et al. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in Southern Brazil. J Med Virol. 2007;79:1455-63.. Considering that the sequences that were classified by the method by YAGYU et al. as subtype D were later confirmed as subtype C by sequencing analysis and knowing that subtype D is extremely rare in the Brazilian population, we expect that the 16 individuals who were classified as having HIV-1 subtype D are actually infected with subtype C virus. If this is true, these data indicate that the subtype C frequency for Itajaí city in the Santa Catarina State for 2006 is 60%. However, the frequencies of subtypes B and C likely include recombinant forms of HIV-1 that cannot be distinguished by the gp41 region that was used in the YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. method. The similarity of the incorrectly classified subtype D and F viruses to the recombinant forms of HIV-1 suggests that the gp41 region that is amplified is too short to clearly distinguish the recombinant forms that are frequently found in the Brazilian population22. Bello G, Guimarães ML, Morgado MG. Evolutionary history of HIV-1 subtype B and F infections in Brazil. AIDS. 2006;20:763-8.,33. Couto-Fernandez JC, Eyer-Silva WA, Guimarães ML, Chequer-Fernandez SL, Grinsztejn B, Delaporte E, et al. Phylogenetic analysis of Brazilian HIV type 1 subtype D strains: tracing the origin of this subtype in Brazil. AIDS Res Hum Retroviruses. 2006;22:207-11.,99. Guimarães ML, dos Santos Moreira A, Loureiro R, Galvão-Castro B, Morgado MG. High frequency of recombinant genomes in HIV type 1 samples from Brazilian southeastern and southern regions. AIDS Res Hum Retroviruses. 2002;18:1261-9.,1919. Sá Filho DJ, Sucupira MC, Caseiro MM, Sabino EC, Diaz RS, Janini LM. Identification of two HIV type 1 circulating recombinant forms in Brazil. AIDS Res Hum Retroviruses. 2006;22:1-13.,2222. Santos AF, Schrago CG, Martinez AM, Mendoza-Sassi R, Silveira J, Sousa TM, et al. Epidemiologic and evolutionary trends of HIV-1 CRF31_BC-related strains in southern Brazil. J Acquir Immune Defic Syndr. 2007;45:328-33.,2424. Santos AF, Sousa TM, Soares EA, Sanabani S, Martinez AM, Sprinz E, et al. Characterization of a new circulating recombinant form comprising HIV-1 subtypes C and B in southern Brazil. AIDS. 2006;20:2011-9.. Analyses of the sequences from the Brazilian recombinants CRF28_BF, CRF29_BF, and CRF31_BC have demonstrated that the gp41 region in the BF recombinant forms will produce a subtype B amplification fragment. The BC recombinant form will produce a subtype C amplification fragment. This clearly demonstrates a limitation of the YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. method for subtype analysis in the Brazilian population.
Finally, it can be concluded that the method described by YAGYU et al. 2929. Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23. is not effective for classifying HIV-1 subtypes in the Brazilian population because of the failure of the method in detecting some HIV-1 subtype variants and recombinant forms of HIV-1. Furthermore, because the primer binding sites were not conserved, the method is not representative enough to allow subtype-specific amplification of gp41.
We are very grateful to the HIV-1 patients who voluntarily attended the health service of the Secretary of Health of Itajaí and to Dr. Rosálie K. Knoll and the team at the LMI that contributed to this research. We also thank Tiago Tolentino de Souza for technical support.
REFERENCES
-
1Agwale SM, Robbins KE, Odama L, Saekhou A, Zeh C, Edubio A, et al. Development of an env gp41-based heteroduplex mobility assay for rapid human immunodeficiency virus type 1 subtyping. J Clin Microbiol. 2001;39:2110-4.
-
2Bello G, Guimarães ML, Morgado MG. Evolutionary history of HIV-1 subtype B and F infections in Brazil. AIDS. 2006;20:763-8.
-
3Couto-Fernandez JC, Eyer-Silva WA, Guimarães ML, Chequer-Fernandez SL, Grinsztejn B, Delaporte E, et al. Phylogenetic analysis of Brazilian HIV type 1 subtype D strains: tracing the origin of this subtype in Brazil. AIDS Res Hum Retroviruses. 2006;22:207-11.
-
4Costa SM, Schechter M, Shindo N, Vicente AC, Oliveira EF, Pinto ME, et al. Sequence and phylogenetic analysis of glycoprotein 120 of an HIV type 1 variant (GWGR) prevalent in Brazil. AIDS Res Hum Retroviruses. 1995;11:1143-5.
-
5Esté JA, Telenti A. HIV entry inhibitors. Lancet. 2007;370(9581):81-8.
-
6Gadelha SR, Shindo N, Cruz JN, Morgado MG, Galvão-Castro B. Molecular epidemiology of human immunodeficiency virus-1 in the state of Ceará, Northeast, Brazil. Mem Inst Oswaldo Cruz. 2003;98:461-3.
-
7Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651-67.
-
8Guimarães ML, Bastos FI, Telles PR, Galvão-Castro B, Diaz RS, Bongertz V, et al. Retrovirus infections in a sample of injecting drug users in Rio de Janeiro City, Brazil: prevalence of HIV-1 subtypes, and co-infection with HTLV-I/II. J Clin Virol. 2001;21:143-51.
-
9Guimarães ML, dos Santos Moreira A, Loureiro R, Galvão-Castro B, Morgado MG. High frequency of recombinant genomes in HIV type 1 samples from Brazilian southeastern and southern regions. AIDS Res Hum Retroviruses. 2002;18:1261-9.
-
10Heeney JL, Dalgleish AG, Weiss RA. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313(5786):462-6.
-
11Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18:182-92.
-
12Locateli D, Stoco PH, Queiroz AT, Alcântara LC, Ferreira LG, Zanetti CR, et al. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in Southern Brazil. J Med Virol. 2007;79:1455-63.
-
13Martinez AM, Barbosa EF, Ferreira PC, Cardoso FA, Silveira J, Sassi G, et al. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev Soc Bras Med Trop. 2002;35:471-6.
-
14Martinez AM, Hora VP, Santos AL, Mendoza-Sassi R, Von Groll A, Soares EA, et al. Determinants of HIV-1 mother-to-child transmission in Southern Brazil. An Acad Bras Cienc. 2006;78:113-21.
-
15Ministério da Sáude. Brasil. Boletim Epidemiológico AIDS e DST. Brasília; Ministério da Saúde; 2011.
-
16Peeters M. Recombinant HIV sequences: their role in the global epidemic. 2000 HIV Sequence Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2000compendium.html
» http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2000compendium.html -
17Pieniazek D, Chunfu Y, Lal RB. Phylogenetic analysis of gp41 envelope of HIV-1 groups M, N and O strains provides an alternate region for subtype determination. The Human retroviruses and Aids 1998 Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/98compendium.html
» http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/98compendium.html -
18Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55-6.
-
19Sá Filho DJ, Sucupira MC, Caseiro MM, Sabino EC, Diaz RS, Janini LM. Identification of two HIV type 1 circulating recombinant forms in Brazil. AIDS Res Hum Retroviruses. 2006;22:1-13.
-
20Sabino EC, Diaz RS, Brigido LF, Learn GH, Mullins JI, Reingold AL, et al. Distribution of HIV-1 subtypes seen in an AIDS clinic in Sao Paulo City, Brazil. AIDS. 1996;10:1579-84.
-
21Sanders RW, Korber B, Lu M, Berkhout B, Moore JP. Mutational analyses and natural variability of the gp41 ectodomain. 2002 HIV Sequence Compendium. Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2002compendium.html
» http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2002compendium.html -
22Santos AF, Schrago CG, Martinez AM, Mendoza-Sassi R, Silveira J, Sousa TM, et al. Epidemiologic and evolutionary trends of HIV-1 CRF31_BC-related strains in southern Brazil. J Acquir Immune Defic Syndr. 2007;45:328-33.
-
23Santos AF, Soares MA. HIV genetic diversity and drug resistance. Viruses. 2010;2:503-31.
-
24Santos AF, Sousa TM, Soares EA, Sanabani S, Martinez AM, Sprinz E, et al. Characterization of a new circulating recombinant form comprising HIV-1 subtypes C and B in southern Brazil. AIDS. 2006;20:2011-9.
-
25Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol Biol Evol. 2011;28:2731-9.
-
26Teixeira SL, Bastos FI, Telles PR, Hacker MA, Brigido LF, de F Oliveira CA, et al. HIV-1 infection among injection and ex-injection drug users from Rio de Janeiro, Brazil: prevalence, estimated incidence and genetic diversity. J Clin Virol. 2004;31:221-6.
-
27Tupinambás U, Ribeiro FA, Aleixo A, Greco D. Treatment switch guided by HIV-1 genotyping in Brazil. Braz J Infect Dis. 2006;10:82-8.
-
28Yagyu F, Ikeda Y, Ariyoshi K, Sugiura W, Wongkhomthong SA, Masuda M, et al. Differentiation of subtypes B and E of human immunodeficiency virus type 1 by polymerase chain reaction using novel env gene primers. J Virol Methods. 2002;101:11-20.
-
29Yagyu F, Okitsu S, Tanamoto K, Ushijima H. Determination of HIV-1 subtypes (A-D, F, G, CRF01_AE) by PCR in the transmembrane region (gp41) with novel primers. J Med Virol. 2005;76:16-23.
Publication Dates
-
Publication in this collection
Apr 2013
History
-
Received
9 May 2012 -
Accepted
21 Aug 2012