Abstract
Capsicum includes ca. 41 species of chili peppers. In this original report we PCR amplified, cloned, sequenced and characterized the 5S rDNA non-transcribed spacer -NTS- in 23 taxa of nine clades of Capsicum, divergent at geographical origin and fruit and chromosome traits, and compared the NTS features throughout Solanaceae. According to GC content, inner variability and regulatory elements, the NTS organizes into three distinct structural regions; genetic variability at the NTS in Capsicum and related genus clusters into defined taxa hierarchies. Based on the reconstruction of a maximum-likelihood phylogenetic tree and phylogenetic networks, NTS sequences of Capsicum and related taxa grouped into well recognized categories -genus, section, clade, species, variety-. An evolutionary scenario arose from combined genetic and phylogenetic NTS data, in which monophyly and lineage diversification over time of Capsicum are addressed. Our analysis is original to include all domesticated species of Capsicum prevailing in germplasm collections and breeding programs, together with a large group of wild taxa that demanded further genetic characterization. The NTS set up as a double purpose marker in Capsicum, to directly evaluate genetic variability and reconstruct phylogenetic relationships to a broad extent, and constitutes a valuable tool for germplasm characterization and evolutionary studies within Solanaceae.
Key words
chili peppers; genetic variability; molecular double purpose marker; phylogeny; ribosomal NTS
Resumen
Capsicum incluye ca. 41 especies de ajíes. En este trabajo original, el espaciador no-transcrito (NTS) del ADNr 5S fue PCR-amplificado, clonado, secuenciado y caracterizado en 23 taxones de nueve clados de Capsicum, divergentes en origen, fruto y cromosomas, y comparado a lo largo de Solanaceae. El NTS se organiza en tres regiones estructurales distintas de acuerdo a contenido GC, variabilidad y elementos reguladores; la variabilidad genética del NTS en Capsicum y géneros relacionados se agrupó en categorías taxonómicas definidas. Las secuencias NTS de Capsicum y taxa relacionados también se agruparon en categorías reconocidas -género, sección, clado, especie, variedad- durante la reconstrucción de un árbol filogenético de máxima-verosimilitud y diversas redes filogenéticas. De la combinación de datos genéticos y filogenéticos del NTS surge un escenario evolutivo que considera monofilia y diversificación de Capsicum a lo largo del tiempo. Nuestro análisis es original al incluir todas las especies domesticadas de Capsicum, mayoritarias en colecciones y programas, además de un amplio número de ajíes silvestres que demandaban mayor caracterización genética. El NTS constituye un marcador de doble propósito en Capsicum, al evaluar directamente variabilidad genética y reconstruir relaciones filogenéticas extensas, además de ser útil a la caracterización de germoplasma y estudios evolutivos en Solanaceae.
Palabras clave
ajíes; variabilidad genética; marcador molecular de doble propósito; filogenia
Introduction
Capsicum is a small genus of Solanaceae with ca. 41 species native to tropical and temperate regions of America, distributing from Mexico to Argentina (Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
; Barboza et al. 2019Barboza GE, Carrizo García C, Leiva González S, Scaldafferro M & Reyes X (2019) Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS One 14: e0209792. DOI: 10.1371/journal.pone.0209792
https://doi.org/10.1371/journal.pone.020...
, 2020Barboza GE, de Bem Bianchetti L & Stehmann JR (2020) Capsicum carassense (Solanaceae), a new species from the Brazilian Atlantic Forest. PhytoKeys 140: 12-138. DOI: 10.3897/phytokeys. 140.47071
https://doi.org/10.3897/phytokeys. 140.4...
). The most important feature in Capsicum is fruit pungency through varying degrees including the sweet and hot chili peppers consumed as vegetables or spices, respectively (Moscone et al. 2007Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE & Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Horticulturae 745: 137-169. DOI: 10.17660/ActaHortic.2007.745.5
https://doi.org/10.17660/ActaHortic.2007...
; Stewart et al. 2007Stewart C, Mazourek M, Stellari GM, O’Connell M & Jahn M (2007) Genetic control of pungency in C. chinense via the Pun1 locus. Journal of Experimental Botany 58: 979-991. DOI: 10.1093/jxb/erl243
https://doi.org/10.1093/jxb/erl243...
). The impact of chili peppers in economy is illustrated by the increasing global production and cultured area (Jarret et al. 2019Jarret RL, Barboza GE, Batista FRC, Berke T, Chou Y-Y, Hulse-Kemp A, Ochoa-Alejo N, Tripodi P, Veres A, Carrizo Garcia C, Csillery G, Huang Y-K, Kiss E, Kovacs Z, Kondrak M, Arce-Rodriguez ML, Scaldaferro MA & Szoke A (2019) Capsicum - an abbreviated compendium. Journal of the American Society for Horticultural Science 144: 3-22. DOI: 10.21273/JASHS04446-18
https://doi.org/10.21273/JASHS04446-18...
; Tripodi & Kumar 2019Tripodi P & Kumar S (2019) The Capsicum crop: an introduction. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 1-8. DOI: 10.1007/978-3-319-97217-6). Most economically important species of chili peppers belong to the Annuum clade -C. annuum, C. chinense, C. frutescens-, cultivated and consumed worldwide, added to C. baccatum, C. chacoense, C. eximium and C. pubescens, mostly appreciated in small markets of South America.
Variability across Capsicum is large, with extensive morphological differences, mainly related to fruit shape, color, and size added to variation in fruit pungency (Walsh & Hoot 2001Walsh BM & Hoot SB (2001) Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear waxy introns. International Journal of Plant Sciences 162: 1409-1418. DOI: 10.1086/323273
https://doi.org/10.1086/323273...
; Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
; Cardoso et al. 2018Cardoso R, Ruas CF, Giacomin RM, Ruas PM, Ruas EA, Barbieri RL, Rodrigues R & Gonçalves LSA (2018) Genetic variability in Brazilian Capsicum baccatum germplasm collection assessed by morphological fruit traits and AFLP markers. PLoS ONE 13: e0196468. DOI: 10.1371/journal.pone.0196468
https://doi.org/10.1371/journal.pone.019...
; Colonna et al. 2019Colonna V, DʼAgostino N, Garrison E, Albrechtsen A, Meisner J, Facchiano A, Cardi T & Tripodi P (2019) Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Scientific Reports 9: 10067. DOI: 10.1038/s41598-019-46136-5
https://doi.org/10.1038/s41598-019-46136...
). Additional variability in the genus is reflected by marked genome size and karyotype divergences, this last useful to delimitate wild and cultivated taxa (Moscone et al. 2007Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE & Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Horticulturae 745: 137-169. DOI: 10.17660/ActaHortic.2007.745.5
https://doi.org/10.17660/ActaHortic.2007...
; Grabiele et al. 2018Grabiele M, Debat HJ, Scaldaferro MA, Aguilera PM, Moscone EA, Seijo GJ & Ducasse DA (2018) Highly GC-rich heterochromatin in chili peppers (Capsicum-Solanaceae): a cytogenetic and molecular characterization. Scientia Horticulturae 238: 391-399. DOI: 10.1016/j.scienta.2018.04.060
https://doi.org/10.1016/j.scienta.2018.0...
; Scaldaferro & Moscone 2019Scaldaferro MA & Moscone EA (2019) Cytology and DNA content variation of Capsicum genomes. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 57-84. DOI: 10.1007/978-3-319-97217-6), but of uncertain phylogenetic weight. Recently, genetic variability and delimitation in Capsicum were also evaluated by distances at molecular level, i.e. by repetitive markers such as microsatellites and ribosomal DNA (Sun et al. 2014aSun Y-L, Choi I-L, Lee Y-B, Choi KY, Hong S-K & Kang H-M (2014a) Molecular diversity and phylogentic analysis of Capsicum annuum varieties using the nrDNA ITS region. Scientia Horticulturae 165: 336-343. DOI: 10.1016/j.scienta.2013.11.009
https://doi.org/10.1016/j.scienta.2013.1...
; Ibarra-Torres et al. 2015Ibarra-Torres P, Valadez-Moctezuma E, Pérez-Grajales M, Rodríguez-Campos J & Jaramillo-Flores ME (2015) Inter- and intraspecific differentiation of Capsicum annuum and Capsicum pubescens using ISSR and SSR markers. Scientia Horticulturae 181: 137-146. DOI: 10.1016/j.scienta.2014.10.054
https://doi.org/10.1016/j.scienta.2014.1...
; Rivera et al. 2016Rivera A, Monteagudo AB, Igartua E, Taboada A, García-Ulloa A, Pomar F, Riveiro-Leira M & Silvar C (2016) Assessing genetic and phenotypic diversity in pepper (Capsicum annuum L.) landraces from North-West Spain. Scientia Horticulturae 203: 1-11. DOI: 10.1016/j.scienta.2016.03.006
https://doi.org/10.1016/j.scienta.2016.0...
). Regarding chili peppers classification, the most comprehensive is the recent evolutionary approach of Carrizo García et al. (2016)Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
. Based on a phylogenetic reconstruction through genetic markers, these authors grouped Capsicum species into eleven well supported clades and discussed the phylogenetic relevance of typical non molecular markers to circumscribe chili peppers.
Currently, more than 99% of materials of major banks belong to the five domesticated Capsicum, restraining the use of wild taxa in improvement programs (Barchenger et al. 2019Barchenger DW, Naresh P & Kumar S (2019) Genetic resources of Capsicum. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 9-23. DOI: 10.1007/978-3-319-97217-6; Jarret et al. 2019Jarret RL, Barboza GE, Batista FRC, Berke T, Chou Y-Y, Hulse-Kemp A, Ochoa-Alejo N, Tripodi P, Veres A, Carrizo Garcia C, Csillery G, Huang Y-K, Kiss E, Kovacs Z, Kondrak M, Arce-Rodriguez ML, Scaldaferro MA & Szoke A (2019) Capsicum - an abbreviated compendium. Journal of the American Society for Horticultural Science 144: 3-22. DOI: 10.21273/JASHS04446-18
https://doi.org/10.21273/JASHS04446-18...
). Horticulture can benefit of the enclosed variability in wild chili peppers, however the characterization of their essential biological, genetic and agronomic features has to be expanded. To germplasm management and breeding purposes, characterization of genetic variability and evolutionary relationships is fundamental. In this sense, a genetic marker that can consider both would be major.
In plant genomes, the ribosomal 5S unit organizes in tandem repeats of multiple copies, at single or various loci, with each copy comprising a gene of 120 nucleotides (nt) and a contiguous non transcribed spacer (NTS) of 100–900 nt (Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
). The gene sequence is vital to life and holds cis regulatory elements to its own transcription (Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
; Szymanski et al. 2003Szymanski M, Barciszewska MZ, Erdmann VA & Barciszewski J (2003) 5S rRNA: structure and interactions. Biochemical Journal 371: 641-651. DOI: 10.1042/BJ20020872
https://doi.org/10.1042/BJ20020872...
), thus it is conserved throughout kingdoms and its low substitution rate allows to reconstruct phylogenetic relationships among distantly related organisms (Hori et al. 1985Hori H, Lim B-L & Osawa S (1985) Evolution of green plants as deduced from 5S rRNA sequences. Proceedings of the National Academy of Sciences of the United States of America 82: 820-823. DOI: 10.1073/pnas.82.3.820
https://doi.org/10.1073/pnas.82.3.820...
; Hori & Osawa 1987Hori H & Osawa S (1987) Origin and evolution of organisms as deduced from 5S ribosomal RNA sequences. Molecular Biology and Evolution 4: 445-472. DOI: 10.1093/oxfordjournals.molbev.a040455
https://doi.org/10.1093/oxfordjournals.m...
). On the contrary, the NTS is more variable and evolves faster in particular at middle region, rather than 5’ and 3’ regions which hold tentative motifs related to transcription of flanking gene sequences (Besendorfer et al. 2005Besendorfer V, Krajacic-Sokol I, Jelenie S, Puizina J, Mlinarec J, Sviben T & Papes D (2005) Two classes of 5S rDNA unit arrays of the silver fir, Abies alba Mill.: structure, organization and evolution. Theoretical and Applied Genetics 110: 730-740. DOI: 10.1007/s00122-004-1899-y
https://doi.org/10.1007/s00122-004-1899-...
). Hence, the NTS sequence demonstrated useful to do phylogenetic inferences at such dispair organisms as Triticeae, Crustaceae or Molluska (Kellogg & Appels 1995Kellogg EA & Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140: 325-343.; Allaby & Brown 2001Allaby RG & Brown TA (2001) Network analysis provides insights into evolution of 5S rDNA arrays in Triticum and Aegilops. Genetics 157: 1331-1341.; Perina et al. 2011Perina A, Seoane D, González-Tizón AM, Rodríguez-Fariña F & Martínez-Lage A (2011) Molecular organization and phylogenetic analysis of 5S rDNA in crustaceans of the genus Pollicipes reveal birth-and-death evolution and strong purifying selection. Evolutionary Biology 11: 304. DOI: 10.1186/1471-2148-11-304
https://doi.org/10.1186/1471-2148-11-304...
; Vizoso et al. 2011Vizoso M, Vierna J, González-Tizón AM & Martínez-Lage A (2011) The 5S rDNA gene family in mollusks: characterization of transcriptional regulatory regions, prediction of secondary structures, and long-term evolution, with special attention to Mytilidae mussels. Journal of Heredity 102: 433-447. DOI: 10.1093/jhered/esr046
https://doi.org/10.1093/jhered/esr046...
). Regarding Solanaceae, the characterization of the NTS structure and evolution proved sufficient to evaluate genetic variability and reconstruct phylogenetic relationships in Nicotiana (Kitamura et al. 2001Kitamura S, Inoue M, Shikazono N & Tanaka A (2001) Relationships among Nicotiana species revealed by the 5S rDNA spacer sequence and fluorescence in situ hybridization. Theoretical and Applied Genetics 103: 678-686. DOI: 10.1007/s001220100643
https://doi.org/10.1007/s001220100643...
; Fulnecek et al. 2002Fulnecek J, Lim KY, Leitch AR, Kovarik A & Matyasek R (2002) Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88: 19-25. DOI: 10.1038/sj.hdy.6800001
https://doi.org/10.1038/sj.hdy.6800001...
; Matyasek et al. 2002Matyasek R, Fulnecek J, Lim KY, Leitch AR & Kovarik A (2002) Evolution of 5S rDNA unit arrays in the plant genus Nicotiana (Solanaceae). Genome 45: 556-562. DOI: 10.1139/g02-017
https://doi.org/10.1139/g02-017...
; Clarkson et al. 2005Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S & Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytologist 168: 241-252. DOI: 10.1111/j.1469-8137.2005.01480.x
https://doi.org/10.1111/j.1469-8137.2005...
) and Solanum section petota (Volkov et al. 2001Volkov RA, Zanke C, Panchuk II & Hemleben V (2001) Molecular evolution of 5S rDNA of Solanum species (sect. Petota): application for molecular phylogeny and breeding. Theoretical and Applied Genetics 103: 1273-1282. DOI: 10.1007/s001220100670
https://doi.org/10.1007/s001220100670...
). In addition, the categorization of tomato varieties via a comparative alignment of gene and NTS sequences was also reported (Sun et al. 2014bSun YL, Kang HM, Kim YS, Baek JP, Zheng SL, Xiang J-J & Hong S-K (2014b) Tomato (Solanum lycopersicum) variety discrimination and hybridization analysis based on the 5S rRNA region. Biotechnology and Biotechnological Equipment 28: 431-437. DOI: 10.1080/13102818.2014.928499
https://doi.org/10.1080/13102818.2014.92...
). As to Capsicum, fluorescent in situ hybridization revealed that the ribosomal 5S gene comprises thousand copies per genome (Kwon & Kim 2009Kwon J-K & Kim B-D (2009) Localization of 5S and 25S rRNA genes on somatic and meiotic chromosomes in Capsicum species of chili pepper. Molecules and Cells 27: 205-209. DOI: 10.1007/s10059-009-0025-z
https://doi.org/10.1007/s10059-009-0025-...
) and persistently map at a single intercalar locus on the short arm of a metacentric chromosome pair (Aguilera et al. 2016Aguilera PM, Debat HJ, Scaldaferro MA, Martí DA & Grabiele M (2016) FISH-mapping of the 5S rDNA locus in chili peppers (Capsicum-Solanaceae). Anais da Academia Brasileira de Ciências 88: 117-125. DOI: 10.1590/0001-37652301620140616
https://doi.org/10.1590/0001-37652301620...
). A preliminary and fundamental report depicted the initial characterization of the 5S rDNA nucleotide sequences in the five domesticated chili peppers (Park et al. 2000Park YK, Park KC, Park CH & Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Molecules and Cells 10: 18-24.). Nevertheless, reference genomes in the genus did not deal with the 5S rDNA (Kim et al. 2014Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, Seo E, Choi J, Cheong K, Kim K-T, Jung K, Lee G-W, Oh S-K, Bae C, Kim S-B, Lee H-Y, Kim S-Y, Kim M-S, Kang B-C, Jo YD, Yang H-B, Jeong H-J, Kang W-H, Kwon J-K, Shin C, Lim JY, Park JH, Huh JH, Kim J-S, Kim B-D, Cohen O, Paran I, Suh MC, Lee SB, Kim Y-K, Shin Y, Noh S-J, Park J, Seo YS, Kwon S-Y, Kim HA, Park JM, Kim H-J, Choi S-B, Bosland PW, Reeves G, Jo S-H, Lee B-W, Cho H-T, Choi H-S, Lee M-S, Yu Y, Choi YD, Park B-S, van Deynze A, Ashrafi H, Hill T, Kim WT, Pai H-S, Ahn HK, Yeam I, Giovannoni JJ, Rose JKC, Sørensen I, Lee S-J, Kim RW, Choi I-Y, Choi B-S, Lim J-S, Lee Y-H & Choi D (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics 46: 270-278. DOI: 10.1038/ng.2877
https://doi.org/10.1038/ng.2877...
, 2017Kim S, Park J, Yeom S-I, Kim Y-M, Seo E, Kim K-T, Kim M-S, Lee JM, Cheong K, Shin H-S, Kim S-B, Han K, Lee J, Park M, Lee H-A, Lee H-Y, Lee Y, Oh S, Lee JH, Choi E, Choi E, Lee SE, Jeon J, Kim H, Choi G, Song H, Lee JK, Lee S-C, Kwon J-K, Lee H-Y, Koo N, Hong Y, Kim RW, Kang W-H, Huh JH, Kang B-C, Yang T-J, Lee Y-H, Bennetzen JL & Choi D (2017) New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biology 18: 210. DOI: 10.1186/s13059-017-1341-9
https://doi.org/10.1186/s13059-017-1341-...
; Qin et al. 2014Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y, Yang Y, Wu Z, Mao L, Wu H, Ling-Hu C, Zhou H, Lin H, González-Morales S, Trejo-Saavedra DL, Tian H, Tang X, Zhao M, Huang Z, Zhou A, Yao X, Cui J, Li W, Chen Z, Feng Y, Niu Y, Bi S, Yang X, Li W, Cai H, Luo X, Montes-Hernández S, Leyva-González MA, Xiong Z, He X, Bai L, Tan S, Tang X, Liu D, Liu J, Zhang S, Chen M, Zhang L, Zhang L, Zhang Y, Liao W, Zhang Y, Wang M, Lv X, Wen B, Liu H, Luan H, Zhang Y, Yang S, Wang X, Xu J, Li X, Li S, Wang J, Palloix A, Bosland PW, Li Y, Krogh A, Rivera-Bustamante RF, Herrera-Estrella L, Yin Y, Yu J, Hu K & Zhang Z (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceedings of the National Academy of Sciences of the United States of America 111: 5135-5140. DOI: 10.1073/pnas.1400975111
https://doi.org/10.1073/pnas.1400975111...
; Ahn et al. 2018Ahn Y-K, Manivannan A, Karna S, Jun T-H, Yang E-Y, Choi S, Kim J-H, Kim D-S & Lee E-S (2018) Whole genome resequencing of Capsicum baccatum and Capsicum annuum to discover single nucleotide polymorphism related to powdery mildew resistance. Scientific Reports 8: 5188. DOI: 10.1038/s41598-018-23279-5
https://doi.org/10.1038/s41598-018-23279...
) and a global characterization of the NTS structure and evolution is still lacking in wild taxa and Capsicum as a whole.
In this original report we examine in depth the nucleotide sequence of the 5S rDNA NTS in Capsicum and overall Solanaceae, and discuss the utility of the NTS to evaluate genetic variability and reconstruct phylogenetic relationships to a broad extent in chili peppers. For this purpose, we PCR amplified, cloned, sequenced and characterized the NTS in 23 wild and cultivated taxa of nine major clades of Capsicum, also divergent at geographical origin and fruit and chromosome traits.
Materials and Methods
Plant materials
Twenty three taxa of Capsicum -including twenty species, five varieties and three cultivars- and Lycianthes rantonnei -sister group- used in this study were identified by Dr. Gloria E. Barboza (Instituto Multidisiplinario de Biología Vegetal, IMBIV, Córdoba, Argentina) and their respective names, clade belonging to, provenance, voucher specimen, herbarium, status, fruit traits and chromosome numbers are detailed in Table 1.
Summary of the material of Capsicum and Lycianthes used in this study in addition to the amplification strategy and public accession of the 5S rDNA sequences.
Methods Isolation, cloning and sequencing of the 5S rDNA
Total DNA was isolated and purified from fresh leaves by CTAB method (Rogers & Bendich 1994Rogers SO & Bendich AJ (1994) Extraction of total cellular DNA from plant, algae and fungi. In: Stanton BG & Schilperoort RA (eds.) A plant molecular biololgy manual. Kluwer Academic Publisher, Dordrecht. Pp. D1/1-8.). Additionally, a phenol chloroform purification prior to an ethanol precipitation were included (Sambrook & Russell 2001Sambrook J & Russell DW (2001) Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 2100p.). DNA was assessed for quality by agarose gel electrophoresis and quantified by spectrophotometry. Nuclear 5S rDNA gene and NTS were PCR amplified using primers derived from conserved regions at the gene, P1 5’GATCCCATCAGAACTCC3’ and P2 5’GGTGCTTTAGTGCTGGTAT3’ (Park et al. 2000Park YK, Park KC, Park CH & Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Molecules and Cells 10: 18-24.) and RT1 5’GGATGCGATCATACCAGC3’ and RT2 5’GAGGGATGCAACACGAGG3’ (Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
). PCR experiments were planned to cover the entire span of the NTS unit (Fig. 1a). In the PCR reactions, the Taq DNA Polymerase “Sequencing Grade” (Promega, USA) was used (1 unit of polymerase; 5 ng of template DNA; 0.5 pmoles of each primer; 200 mM of dNTPs; 5 µl of 10X buffer) and 36 cycles (94 °C 1 min, 57 °C 1 min, 72 °C 1 min) with a final extension at 72 °C 5 min were performed. PCR products were electrophoresed in 1.4% agarose (Fig. 1b), gel isolated, purified by the GFX kit (Amersham Pharmacia, USA), cloned in pCR2.1 TOPO and transformed into “TOP10 One Shot” E. coli (Invitrogen, USA) according to manufacturer instructions. Clones were subsequently grown in LB media with ampicillin and the obtained cultures were subjected to plasmidic DNA minipreparations using the Wizard Plus Minipreps DNA Purification System (Promega, USA). Plasmidic DNAs were then digested with EcoRI (NEB, USA) according to manufacturer instructions and visualized in 1% agarose gel to check the stability of the inserts. Selected clones were bidirectionally Sanger sequenced at Macrogen (Korea) and BLASTN searches against the nr/nt nucleotide collection were conducted with the resulting sequences to confirm their identities prior to further analysis. Sequences -and their major annotated features (see below)- were deposited at DDBJ/ENA/GenBank (Sayers et al. 2019Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, Holmes JB, Kim S, Kimchi A, Kitts PA, Lathrop S, Lu Z, Madden TL, Marchler-Bauer A, Phan L, Schneider VA, Schoch CL, Pruitt KD & Ostell J (2019) Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 47: D23-D28. DOI: 10.1093/nar/gky1069
https://doi.org/10.1093/nar/gky1069...
) under the accessions MK650892-MK651009 and JF773766 (Capsicum), and MK638982-MK638984 (L. rantonnei). A summary of the amplification strategy and public accession of the 5S rDNA sequences is presented in Table 1.
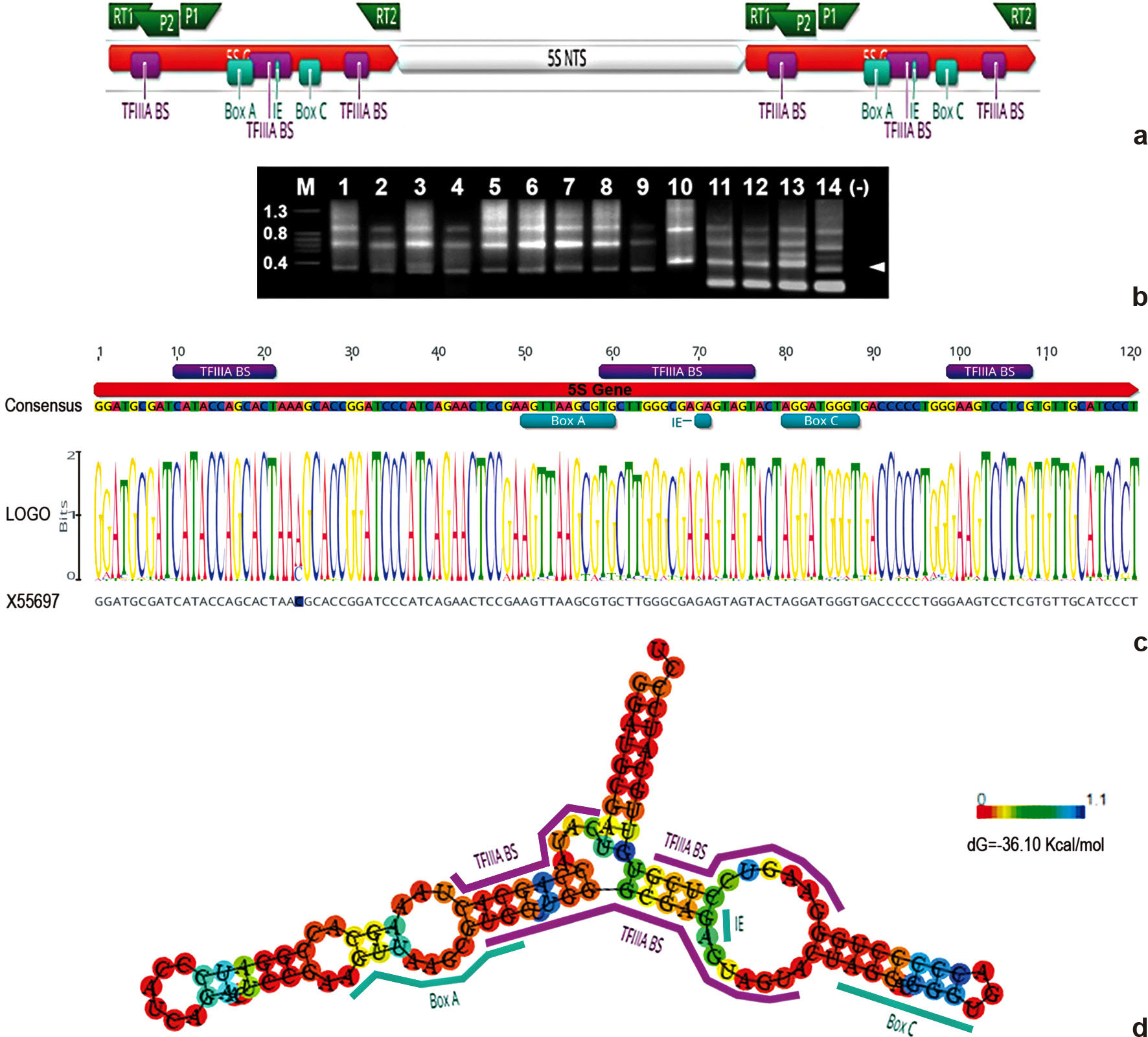
a. Amplification strategy of the 5S NTS in Capsicum via P1/P2 and RT1/RT2 primer pairs; TFIIIA BS = TFIIIA transcript binding site (Kellogg & Appels 1995Kellogg EA & Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140: 325-343.); Box A, Box C and IE = internal control regions (Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301... ). b. PCR products of 5S regions; (P1/P2) 1 = C. flexuosum; 2 = C. praetermissum; 3 = C. baccatum var. umbilicatum; 4 = C. baccatum var. baccatum; 5 = C. annuum var. annuum; 6 = C. annuum var. glabriusculum; 7 = C. frutescens; 8 = C. chinense; 9 = C. chacoense; 10 = C. tovarii; (RT1/RT2) 11 = C. hunzikerianum; 12 = C. schottianum; 13 = C. campylopodium; 14 = C. coccineum; note the typical amplification ladder pattern of the repetitive 5S region; arrowhead point out to purified products; M, marker (Kbp); (-), negative control. c. Consensus and LOGO 5S gene sequences in Capsicum. d. Secondary structure of the consensus 5S gene in Capsicum displaying the usual eukaryote mode (Szymanski et al. 2003Szymanski M, Barciszewska MZ, Erdmann VA & Barciszewski J (2003) 5S rRNA: structure and interactions. Biochemical Journal 371: 641-651. DOI: 10.1042/BJ20020872
https://doi.org/10.1042/BJ20020872... ); red to blue colours scale correspond to high to low probabilities, respectively.
Sequence analysis
The editing, multiple alignments and annotation of the nucleotide sequences were performed in Geneious Pro 11.0.5 (Biomatters Ltd.). The 5S gene was annotated following preceding criteria (Kellogg & Appels 1995Kellogg EA & Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140: 325-343.; Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
), pointing out the TFIIIA transcript binding sites and the internal control regions (Box A, Box C and IE), respectively. Capsicum 5S gene majority consensus sequence was obtained through reference mapping of gene sequences onto tomato X55697. Secondary structure analysis of the consensus gene was performed at the RNAfold server (<http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi>). PolIII transcription termination site and other putative regulatory elements at the NTS were annotated according to preceding criteria (Venkateswarlu et al. 1991Venkateswarlu K, Lee SW & Nazar RN (1991) Conserved upstream sequence elements in plant 5S ribosomal RNA-encoding genes. Gene 105: 249-253.; Drouin & Moniz de Sá 1995Drouin G & Moniz de Sá M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Molecular Biology and Evolution 12: 481-493. DOI: 10.1093/oxfordjournals.molbev.a040223
https://doi.org/10.1093/oxfordjournals.m...
; Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
). Whole annotated sequences are available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1> as supplementary material (Suppl.) in Mendeley Data as Capsicum_NTS.gff and Lycianthes_NTS.gff files. Overall Solanaceae NTS sequences at the DDBJ/ENA/GenBank were retrieved from NCBI and further characterized as described (Suppl.).
Phylogenetic analysis
An alignment matrix of 553 characters comprising 255 entire length NTS sequences of Capsicum and related taxa with comparable NTS information -Lycianthes, Solanum, Datura, Atropa- was constructed (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 12). MAFFT v.7.388 (Kazutaka & Standley 2013Kazutaka K & Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772-780. DOI: 10.1093/molbev/mst010
https://doi.org/10.1093/molbev/mst010...
) multiple sequence alignment was performed via the E-INS-i algorithm, according to multiple conserved domains and long gaps sequences with scoring matrix from Kimura’s two parameter model, and then corrected by eye inspection.
Phylogenetic relationships were inferred via FastTree 2.1.5 -set at Geneious Pro 11.0.5- using the MAFFT NTS alignment matrix; this software infers approximately maximum likelihood (ML) phylogenetic trees, uses heuristic Neighbor Joining, reduces the length of the tree by NNIs and SPRs, maximizes the tree’s likelihood with NNIs and finally estimates splits reliability by Shimodaira-Hasegawa test and 1,000 default resamples (local support values: 0–1) (Price et al. 2010Price MN, Dehal PS & Arkin AP (2010) FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. DOI: 10.1371/journal.pone.0009490
https://doi.org/10.1371/journal.pone.000...
). The substitution model General Time Reversible (GTR) with a gamma -20- distribution of rates of evolution among sites was selected after submission of the alignment matrix to MEGA 7.0.26 (Kumar et al. 2015Kumar S, Stecher G & Tamura K (2015) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870-1874. DOI: 10.1093/molbev/msw054
https://doi.org/10.1093/molbev/msw054...
) to find best model via Bayesian information criterion (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Tab. 4). Pseudocounts criterion was selected according to the highly gapped alignment matrix, so as to ML and minimum evolution rounds. Furthermore, a phylogenetic network of sequences was constructed via SplitsTree4 (Huson & Bryant 2006Huson DH & Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254-267. DOI: 10.1093/molbev/msj030
https://doi.org/10.1093/molbev/msj030...
) employing the NTS alignment matrix, the Neighbornet algorithm, the uncorrected P distance and 100 bootstrap replicates. Finally, nucleotide substitution rates at the NTS were calculated after submission of the alignment matrix to MEGA 7.0.26 in which nucleotide diversity (π) estimates of phylogenetically relevant groups were obtained -substitution model K2+G; 100 bootstrap replicates- and by considering evolutionary divergence times -million years ago, MYA- in Solanaceae. For this purpose we followed the approach of Sarkinen et al. (2013)Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
https://doi.org/10.1186/1471-2148-13-214...
, in which splits times are unambigously stated on the tree, instead of the recalibrated times for Solanaceae of De-Silva et al. (2017)De-Silva DL, Mota LL, Chazot N, Mallarino R, Silva Brandão KL, Gómez Piñerez LM, Freitas AVL, Lamas G, Joron M, Mallet J, Giraldo CE, Uribe S, Sarkinen T, Knapp S, Jiggins CD, Willmott KR & Elias M (2017) North Andean origin and diversification of the largest ithomiine butterfly genus. Scientific Reports 7: 45966. DOI: 10.1038/srep45966
https://doi.org/10.1038/srep45966...
; both phylogenies share similar node support and topology, but lineages ages are ca. 25% older on average in the most recent analysis (De-Silva et al. 2017De-Silva DL, Mota LL, Chazot N, Mallarino R, Silva Brandão KL, Gómez Piñerez LM, Freitas AVL, Lamas G, Joron M, Mallet J, Giraldo CE, Uribe S, Sarkinen T, Knapp S, Jiggins CD, Willmott KR & Elias M (2017) North Andean origin and diversification of the largest ithomiine butterfly genus. Scientific Reports 7: 45966. DOI: 10.1038/srep45966
https://doi.org/10.1038/srep45966...
). Nomenclature on clades in Capsicum follows preceding criteria (Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
).
Results and Discussion
Molecular characterization of the NTS: genetic variability in Capsicum and Solanaceae
The entire span of the NTS unit of the 5S rDNA gene was sequenced in twenty three taxa of Capsicum and L. rantonnei, comprising three to eight paralog copies for each taxon (Tab. 1; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Tab. 1). Primary and secondary structures of the 5S gene of Capsicum are also shown (Fig. 1c,d). An exhaustive multiple alignment of the 126 NTS sequences -including previous data (Park et al. 2000Park YK, Park KC, Park CH & Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Molecules and Cells 10: 18-24.)- (Fig. 2) allowed to recognize three major structural regions (SRI-III) according to nucleotide composition, inner variability and regulatory elements related to transcription of upstream and downstream 5S gene. In addition, main structural features were identified that cluster into well known clades of Capsicum (Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
). A summary of main features at the NTS is shown in Table 2, considering individual taxon variability, that is paralog diversity, and that of Capsicum as a whole, through estimates of pairwise identity, mean GC bases content and mean nucleotide lenght for each SR and the full NTS sequence. In addition, a particular ribosomal related sequence (JF773766) containing an AuSINE member of transposable elements detected in the expected NTS was further characterized (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 3).
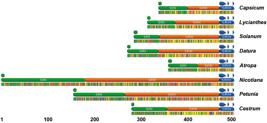
Structural characterization of the 5S NTS in chili peppers. Multiple alignment and annotation of 126 sequences linked to 23 taxa of Capsicum and L. rantonnei. SR = structural regions. Functional features related to transcription of upstream (polyT) and downstream (-30 TATA, -12 GC, -1 C/T) 5S CR are shown. 1-3: L. rantonnei (Sister group); 4-8: C. rhomboideum; 9-12: C. geminifolium; 13-16: C. pereirae; 17-21: C. villosum; 22-25: C. friburgense; 26-33: C. schottianum; 34-37: C. recurvatum; 38-43: C. campylopodium; 44-51: C. hunzikerianum; 52-56: C. flexuosum; 57-62: C. coccineum; 63-68: C. pubescens; 70-73: C. eximium; 74-80: C. tovarii; 81-84: C. chacoense; 85-90: C. praetermissum; 91-93: C. baccatum var. baccatum; 94-97: C. baccatum var. umbilicatum; 98-101: C. baccatum var. pendulum; 103-107: C. frutescens; 109-113: C. chinense; 115-119: C. annuum var. glabriusculum; 120-125: C. annuum var. annuum. 69, 102, 108, 114 and 126 correspond to cultivated taxa under AF217950-4 (Park et al. 2000Park YK, Park KC, Park CH & Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Molecules and Cells 10: 18-24.). Major insertions at C. chacoense, C. tovarii and Andean clade taxa added to sites over 90% gaps are hided. Note main structural features defining clades at Capsicum.
Structural characterization of the 5S NTS in chili peppers. Major insertion segments as useful markers at C. tovarii (123 nt), Andean clade taxa (13 nt) and C. chacoense (7 nt). Note the unusually large SRI of C. tovarii that contains a repeated region at 5’ and 3’ (light green) associated to a region fairly similar to a 5S gene (light red), probably originated by unequal crossing over within the ribosomal region.
The SRI in Capsicum is particularly AT rich (35% mean GC content) and variable in length, ranging from 40.3 to 199.9 nucleotides long (nt), with a mean of 68.8 nt among taxa. SRI of C. tovarii -and also the NTS- is the largest in Capsicum due to a major insertion of 123 nt which consists of a repeated region at 5’ and 3’ associated to a region fairly similar to a 5S gene, probably originated by unequal crossing over within the ribosomal region (Fig. 3; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 12). Capsicum chacoense exhibits an insertion (7 nt) of repetitive nature at a poly(T) stretch (Fig. 3; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 12), a distinctive marker among taxa of Baccatum clade. The poly(T) stretch at 5’ end SRI, tentative PolIII transcription termination site of the 5S gene (Drouin & Moniz de Sá 1995Drouin G & Moniz de Sá M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Molecular Biology and Evolution 12: 481-493. DOI: 10.1093/oxfordjournals.molbev.a040223
https://doi.org/10.1093/oxfordjournals.m...
; Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
), is also variable in length among chili peppers (6–35 nt). This poly(T) sequence is typically 6–7 nt long but unusually larger in members of Tovarii, Baccatum and Annuum clades, which also display a distinctive deletion of 5 nt (TGTCG) downstream this region (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 12). Further, members of Atlantic Forest clade present a major deletion (21 nt), without affecting the poly(T) stretch, at 5’ end of SRI. Meiotic unequal exchange may help to explain the loss of segments in groups above described (Fig. 4). Moreover, all chili peppers, with the exception of the Andean clade taxa, display a CT block at 3’ end SRI and this region is absent in L. rantonnei (Fig. 2).
a-b. Probable scenario on the origin of the current SRI – a. of Atlantic Forest clade members; b. that shared by Tovarii, Baccatum and Annuum clades taxa. 1: simplified depiction of usual homologous chromosomes pairing at the ribosomal region. 2: same region misaligned by repeated sequences suffering an unequal exchange (orange triangles) with loss of segments (Del). 3: resultant SRI.
The SRII in Capsicum is highly GC rich (70.3% mean GC content), averaging 76.2 nt long among taxa, and the most variable in length at the NTS. Hence, this region is ca. 120 nt in members of Andean clade, which display a major insertion (13 nt) of repetitive nature at the middle SRII (Fig. 3; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 12). In contrast, the remarkably short 8 nt SRII of C. coccineum (Bolivian clade) -almost entirely deleted- which contains a unique repeated motif CGGAGG, probably arose by unequal exchange of similar motifs at this region (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 5). In addition, C. flexuosum presents a large deletion of around 28 nt at the middle SRII and C. praetermissum contains a large purine-rich block deletion of 12 nt near the start of its SRII (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Figs. 4; 5). Considering the three varieties of C. baccatum, the central part of the SRII reveal as the utmost divergent (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 1). Further, the middle portion and 3’ end of the SRII are the most variable in length at the NTS of members of Atlantic Forest clade (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 2). At this point, the observed extreme length variation at the SRII in Capsicum agree with the hypothesis that the middle region of the NTS in plants is prone to accumulate more insertions and deletions (indels) than the rest of the spacer, probably associated to the lack of inherent regulatory elements (Besendorfer et al. 2005Besendorfer V, Krajacic-Sokol I, Jelenie S, Puizina J, Mlinarec J, Sviben T & Papes D (2005) Two classes of 5S rDNA unit arrays of the silver fir, Abies alba Mill.: structure, organization and evolution. Theoretical and Applied Genetics 110: 730-740. DOI: 10.1007/s00122-004-1899-y
https://doi.org/10.1007/s00122-004-1899-...
). Particularly, the SRII of the NTS in Capsicum appears to evolve via duplication, deletion and base substitution of CGGGG-like motifs.
Structural characterization of the 5S NTS in Solanaceae. Note the consensus sequences for each taxa and their comparable annotated functional features related to transcription of upstream (poly(T)) and downstream (-30 TATA-like, -12 GC, -1 C/T) 5S gene added to the NTS organization in three structural regions (SRI-III) according to differences on the GC content and inner variability. For details see Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Figs. 7-10.
Finally, the SRIII in Capsicum is distinctly AT rich (35.1% mean GC content) and the most conserved considering length (ca. 30 nt long) and overall pairwise identity (81.6%). On the contrary, overall pairwise identity at the SRI and SRII of the NTS in Capsicum is ca. 50%. The SRIII of chili peppers encloses similar motifs to those found in other flowering plants at the NTS 3’ end, which are suggested to affect PolIII transcription of downstream 5S gene. In this sense, common but less conserved AT rich sequences at ca. -26, assumed to function in the manner of the -10 box of bacterial promoter TATAAT (Reznikoff et al. 1985Reznikoff WS, Siegele DA, Cowing DW & Gross CA (1985) The regulation of transcription initiation in bacteria. Annual Review of Genetics 19: 355-387. DOI: 10.1146/annurev.ge.19.120185.002035
https://doi.org/10.1146/annurev.ge.19.12...
) instead as a typical eukaryote TATA box, were earlier addressed (Venkateswarlu et al. 1991Venkateswarlu K, Lee SW & Nazar RN (1991) Conserved upstream sequence elements in plant 5S ribosomal RNA-encoding genes. Gene 105: 249-253.). Later, the motif TATATA at ca. -30 was recognized in Arabidopsis NTS as necessary for in vitro transcription of the 5S gene (Cloix et al. 2002Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
https://doi.org/10.1101/gr.181301...
). In accordance, around the position -30 in Capsicum NTS, a consensus 92% AT rich stretch of 13 nt -TTTAATAGAATTT- is found, annotated here as TATA like sequence (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 11). This motif is highly conserved throughout examined chili peppers, presenting an overall pairwise identity of 83.7%. Further expected transcription regulatory elements were found downstream the TATA like sequence in Capsicum NTS, i.e. the GC dinucleotide at -12 and a final C at -1 of the transcription initiation site (Fig. 2). In some cases a tetranucleotide motif GCGC is found at -14 (C. friburgense, C. flexuosum, C. eximium) and solely paralog copies of C. pubescens diverge at this region exhibiting GC or GCGC at position -14 (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Figs. 2; 5). Different to a 3’ C at -1, the NTS of some chili peppers -members of Andean, Atlantic Forest, Flexuosum and Bolivian clades- terminate with a T nucleotide (Fig. 2; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Figs. 2; 5; 6) as previously reported for Nicotiana (Fulnecek et al. 2002Fulnecek J, Lim KY, Leitch AR, Kovarik A & Matyasek R (2002) Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88: 19-25. DOI: 10.1038/sj.hdy.6800001
https://doi.org/10.1038/sj.hdy.6800001...
).
To highlight the phylogenetic relevance of overall NTS traits associated to chili pepper identification, 5S ribosomal NTS sequences of Solanaceae available in the literature were revisited in view of the original features found in Capsicum and Lycianthes. To this end, earlier addressed central features from the NTS of Solanum section petota (Volkov et al. 2001Volkov RA, Zanke C, Panchuk II & Hemleben V (2001) Molecular evolution of 5S rDNA of Solanum species (sect. Petota): application for molecular phylogeny and breeding. Theoretical and Applied Genetics 103: 1273-1282. DOI: 10.1007/s001220100670
https://doi.org/10.1007/s001220100670...
), separate sections of Nicotiana (Kitamura et al. 2001Kitamura S, Inoue M, Shikazono N & Tanaka A (2001) Relationships among Nicotiana species revealed by the 5S rDNA spacer sequence and fluorescence in situ hybridization. Theoretical and Applied Genetics 103: 678-686. DOI: 10.1007/s001220100643
https://doi.org/10.1007/s001220100643...
; Fulnecek et al. 2002Fulnecek J, Lim KY, Leitch AR, Kovarik A & Matyasek R (2002) Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88: 19-25. DOI: 10.1038/sj.hdy.6800001
https://doi.org/10.1038/sj.hdy.6800001...
; Matyasek et al. 2002Matyasek R, Fulnecek J, Lim KY, Leitch AR & Kovarik A (2002) Evolution of 5S rDNA unit arrays in the plant genus Nicotiana (Solanaceae). Genome 45: 556-562. DOI: 10.1139/g02-017
https://doi.org/10.1139/g02-017...
; Clarkson et al. 2005Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S & Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytologist 168: 241-252. DOI: 10.1111/j.1469-8137.2005.01480.x
https://doi.org/10.1111/j.1469-8137.2005...
; Fulnecek & Kovarik 2007Fulnecek J & Kovarik A (2007) Low abundant spacer 5S rRNA transcripts are frequently polyadenylated in Nicotiana. Molecular Genetics and Genomics 278: 565-573. DOI: 10.1007/s00438-007-0273-6
https://doi.org/10.1007/s00438-007-0273-...
) and Atropa belladona (Volkov et al. 2017Volkov RA, Panchuk II, Borisjuk NV, Hosiawa-Baranska M, Maluszynska J & Hemleben V (2017) Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biology 17: 21. DOI: 10.1186/s12870-017-0978-6
https://doi.org/10.1186/s12870-017-0978-...
) were reanalyzed together with partially defined NTS of Petunia (Frasch et al. 1989Frasch M, Wenzel W & Hess D (1989) The nucleotide sequences of nuclear 5S rRNA genes and spacer regions of Petunia hybrida. Nucleic Acids Research 17: 2857. DOI: 10.1093/nar/17.7.2857
https://doi.org/10.1093/nar/17.7.2857...
; Venkateswarlu et al. 1991Venkateswarlu K, Lee SW & Nazar RN (1991) Conserved upstream sequence elements in plant 5S ribosomal RNA-encoding genes. Gene 105: 249-253.) and NTS sequences of Cestrum (Sykorova et al. 2003Sykorova E, Lim KY, Fajkus J & Leitch AR (2003) The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 112: 164-172. DOI: 10.1007/s00412-003-0256-2
https://doi.org/10.1007/s00412-003-0256-...
), Datura (Carles et al. 2005Carles M, Cheung MKL, Moganti S, Dong TTX, Tsim KW, Ip NY & Sucher NJ (2005) A DNA microarrey for the authentication of toxic traditional chinese medicinal plants. Planta Medica 71: 580-584. DOI: 10.1055/s-2005-864166
https://doi.org/10.1055/s-2005-864166...
) and Solanum section Lycopersicon (Sun et al. 2014bSun YL, Kang HM, Kim YS, Baek JP, Zheng SL, Xiang J-J & Hong S-K (2014b) Tomato (Solanum lycopersicum) variety discrimination and hybridization analysis based on the 5S rRNA region. Biotechnology and Biotechnological Equipment 28: 431-437. DOI: 10.1080/13102818.2014.928499
https://doi.org/10.1080/13102818.2014.92...
) that lack further characterization. At this regard, detailed multiple alignments of the NTS sequences in those taxa of Solanaceae -Datura, Solanum, Atropa, Nicotiana, Petunia, Cestrum (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Tab. 2, Suppl. Figs. 7-10)- which split from Capsicum and Lycianthes 18 to 30 MYA ago (Sarkinen et al. 2013Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
https://doi.org/10.1186/1471-2148-13-214...
) and their overall comparison, allowed to recognize common and divergent features among nightshades. Our results suggest that Solanaceae NTS typically organizes in three distinct structural regions (SRI-III) according to differences on the GC content, internal variability and regulatory elements associated to transcription of flanking 5S gene (Fig. 5). Estimates of pairwise identity, mean GC bases content and mean nucleotide length for each SR and the full NTS sequence of Solanaceae are summarized in Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Tab. 3. The NTS is variable in length among compared genera, ranging from ca. 138 nt in Atropa to 415.2 nt in Nicotiana, while the NTS of C. coccineum -99 nt- and N. tabacum -527 nt- are the most asymmetrically variable in length. The SRI and SRIII are highly to moderate AT rich, with a mean GC content ranging from 17.6% in Atropa to 37.7% in Lycianthes for the former, and 24.6% in Solanum to 40.0% in Datura for the latter region. On the other hand, the SRII exhibits the highest GC content among taxa, reaching 46.8% in Petunia to 70.3% in chili peppers and varies greatly in length from 8 nt in C. coccineum to 304 nt in N. otophora and N. cordifolia. Contrary to Capsicum, the length and nucleotide composition of the poly(T) stretch are quite constant features all over Solanaceae NTS. Hence, comparable short motifs comprising five to seven nt long are regular, i.e. TTTTT (Cestrum), CTTTTT (Lycianthes, Petunia, Nicotiana), TCTTTT (Datura), CTTTTTT (Atropa) and CCTTTTT (Solanum), as detailed in Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Figs. 7-10. According to the multiple alignment of 311 informative sequences from eight analyzed genera, the SRIII (69.3% AT rich) reveals as the most conserved NTS segment regarding length (ca. 30 nt long) and overall pairwise identity (78.3%) throughout Solanaceae. Comparable structural features at the SRIII considering each genera and full Solanaceae -majority consensus and LOGO sequences- and key functional traits related to transcription of downstream 5S gene, i.e. -30 TATA like, -12 GC, -1 C, are illustrated at Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 11. The consensus SRIII of Solanaceae encompasses a 92% AT rich stretch of 13 nt identical to that of Capsicum -TTTAATAGAATTT-, highly conserved throughout the examined taxa, with Petunia displaying the major deviation from this motif. The dinucleotide motif GC at position -12 is common to three genera while four genera own the tetranucleotide GCGC at -14 and solely Atropa display GC at -14. Finally, at position -1 of the transcription initiation site C or T nucleotides were found, and motifs GAC, GAT and GTC are usual terminal trinucleotides for the NTS in Solanaceae.
At this point, the molecular variability at the NTS demonstrates useful to classify a broad collection of wild and cultivated chili peppers at the specific and clade levels, and even identify related genera of Solanaceae compared among each other, constituting a marker of phylogenetic relevance.
Reconstruction of phylogenetic relationships in Capsicum and related Solanaceae via the NTS
An exhaustive multiple alignment of 255 entire length NTS sequences from Capsicum and related taxa of Solanaceae that hold comparable NTS information -Lycianthes, Solanum, Datura, Atropa- was further achieved via MAFFT and manual curation. The resultant matrix of 553 characters exposed ancient blocks of nucleotides and novel divergent segments among considered taxa (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 12) facilitating the NTS based phylogenetic reconstruction in chili peppers.
To begin, a bootstrap phylogenetic network of the NTS sequences of Capsicum and comparable related taxa in Solanaceae was constructed employing the NTS matrix (Fig. 6). As a first outcome NTS sequences clustered into well defined taxa hierarchies, i.e. genus, section, clade. At this regard, paralog copies in each species of Capsicum also grouped together, with the exception of some related to the Atlantic Forest clade, commented below. Respect to the NTS network at Solanum, section Dulcamara shown sister to sections Petota and Lycopersicon -which include potato and tomato- as expected (clade Potato; Sarkinen et al. 2013Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
https://doi.org/10.1186/1471-2148-13-214...
). Further, Capsicum emerges as monophyletic as in previous analysis (Walsh & Hoot 2001Walsh BM & Hoot SB (2001) Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear waxy introns. International Journal of Plant Sciences 162: 1409-1418. DOI: 10.1086/323273
https://doi.org/10.1086/323273...
; Buso et al. 2002Buso GSC, Amaral ZPS, Bianchetti LDB & Ferreira ME (2002) Análise de seqüências de DNA cloroplástico de espécies do gênero Capsicum. Boletim de Pesquisa e Desenvolvimiento. Vol. 37. Embrapa Recursos Genéticos e Biotecnologia, Brasilia. 15p.; Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
), though its ancient NTS node is right close to Lycianthes. In accordance, Capsicum and Lycianthes hold a complex relationship that justify further consideration (Sarkinen et al. 2013Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
https://doi.org/10.1186/1471-2148-13-214...
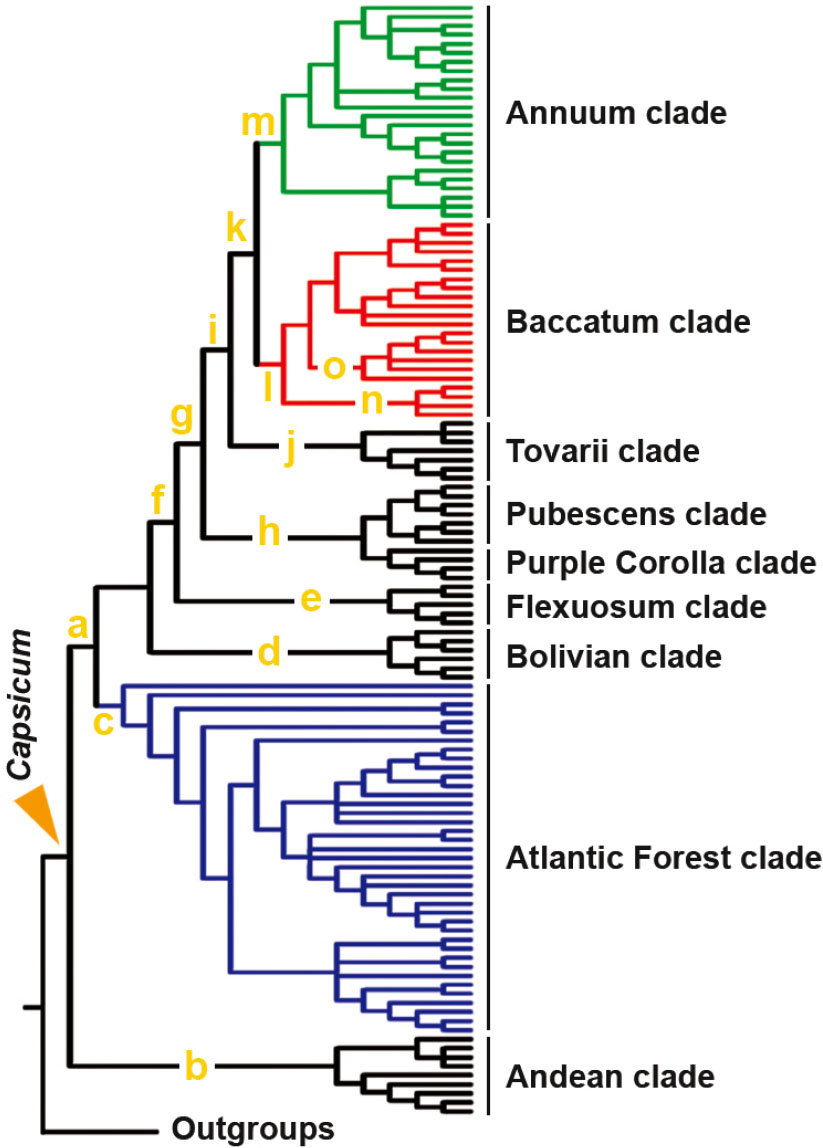
). Among chili peppers, three major lineages are recognized, 1) Andean clade, basal and sister to 2) a superclade comprising Flexuosum, Bolivian and Atlantic Forest clades, and 3) a superclade involving Pubescens and Purple Corolla clades sisters to Tovarii, Baccatum and Annum clades, in which Tovarii is sister to the latter two.
Bootstrap phylogenetic network of the 5S NTS sequences of Capsicum and comparable related taxa -Lycianthes, Solanum, Datura, Atropa-. The network was constructed via SplitsTree4 employing the NTS alignment matrix of Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Fig. 12, the Neighbornet algorithm, the uncorrected P distance and 100 boostrap replicates. Note the major lineages comprising Capsicum. * = C. schottianum paralogs; ** = C. villosum paralogs. Scale bar indicates the scale of the network branches.
Subsequently, a ML phylogenetic tree of the NTS sequences of Capsicum, and those comparable related taxa in the family, was also built via the NTS matrix, displaying acceptable main support values, in a range from moderate to high (0.70–0.99) (Fig. 7; Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 13). As in the NTS network, overall sequences fall into recognized categories, i.e. species, genus, section, clade, except from some paralog copies of Atlantic Forest clade members, next discussed. Topology of outgroups to Capsicum is also equivalent, including that within Solanum, and monophyly of chili peppers is supported too (0.87). In addition, the NTS ML tree shows that the non pungent Andean clade is highly supported (0.91) -and basal as previously reported (Walsh & Hoot 2001Walsh BM & Hoot SB (2001) Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear waxy introns. International Journal of Plant Sciences 162: 1409-1418. DOI: 10.1086/323273
https://doi.org/10.1086/323273...
; Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
)-, so as too the Atlantic Forest clade (0.99) and a super cluster formed by the rest of Capsicum (0.92). Both NTS network and tree support the hypothesis that x = 13 evolved twice and independently in Capsicum, in the Andean clade and in the Atlantic Forest clade of pungent members (Moscone et al. 2007Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE & Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Horticulturae 745: 137-169. DOI: 10.17660/ActaHortic.2007.745.5
https://doi.org/10.17660/ActaHortic.2007...
; Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
). A well supported group (0.99) with identical topology to that of the NTS network is formed by Pubescens, Purple Corolla, Tovarii, Baccatum and Annum clades (P-PC-T-B-A), concordantly to earlier results of Carrizo García et al. (2016)Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
, solely that here C. pubescens and C. eximium appear clustered with high support (0.98) and displaying observable synapomorphies (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 5). At this regard, on their phylogenetic analysis in Capsicum from AFLP data, Ibiza et al. (2012)Ibiza VP, Blanca J, Cañizares J & Nuez F (2012) Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genetic Resources and Crop Evolution 59: 1077-1088. DOI: 10.1007/s10722-011-9744-z
https://doi.org/10.1007/s10722-011-9744-...
also reported a high support grouping (85% bootstrap) of C. pubescens and C. eximium. The well supported phylogenetic group P-PC-T-B-A is based on x = 12 and includes to the main consumed chili peppers worldwide -C. annuum, C. chinense, C. frutescens- and in South America -C. baccatum, C. chacoense, C. eximium, C. pubescens- that can cross hybridize to diverse extent (Walsh & Hoot 2001Walsh BM & Hoot SB (2001) Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear waxy introns. International Journal of Plant Sciences 162: 1409-1418. DOI: 10.1086/323273
https://doi.org/10.1086/323273...
; Onus & Pickersgill 2004Onus AN & Pickersgill B (2004) Unilateral incompatibility in Capsicum (Solanaceae): occurrence and taxonomic distribution. Annals of Botany 94: 289-295. DOI: 10.1093/aob/mch139
https://doi.org/10.1093/aob/mch139...
), and constitute the most valuable genetic resources in germplasm banks and breeding programs (Jarret et al. 2019Jarret RL, Barboza GE, Batista FRC, Berke T, Chou Y-Y, Hulse-Kemp A, Ochoa-Alejo N, Tripodi P, Veres A, Carrizo Garcia C, Csillery G, Huang Y-K, Kiss E, Kovacs Z, Kondrak M, Arce-Rodriguez ML, Scaldaferro MA & Szoke A (2019) Capsicum - an abbreviated compendium. Journal of the American Society for Horticultural Science 144: 3-22. DOI: 10.21273/JASHS04446-18
https://doi.org/10.21273/JASHS04446-18...
; Tripodi & Kumar 2019Tripodi P & Kumar S (2019) The Capsicum crop: an introduction. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 1-8. DOI: 10.1007/978-3-319-97217-6). On the other hand, the placement of the variable pungent C. flexuosum as sister of P-PC-T-B-A group is moderately sustained (0.70) in the NTS ML tree. At this point, it is worth noting that the phylogenetic positions of Flexuosum and Bolivian clades with respect to the Atlantic Forest clade and the P-PC-T-B-A cluster differ considering the NTS tree or network approaches (Fig. 6 vs. 7). In this sense, similar incongruences were also pinpointed before on the DNA based phylogenetic reconstruction of Capsicum considering maximum parsimony or bayesian analysis (Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
). The true phylogenetic position of C. flexuosum -x = 12- and C. coccineum -x = ?- is a relevant tool to future breeding programs aiming to introduce useful genetic traits to main consumed chili peppers based on x = 12, directly or even via x = 13 Atlantic Forest clade members intermediates, if it is possible. Currently, studies on the reproductive biology of C. flexuosum and C. coccineum are lacking or incipient (Carrizo García 2011Carrizo García C (2011) Fruit characteristics, seed production and pollen tube growth in the wild chilli pepper Capsicum flexuosum. Flora 206: 334-340. DOI: 10.1016/j.flora.2010.05.008
https://doi.org/10.1016/j.flora.2010.05....
). Regarding C. tovarii, the moderate support (0.74) as sister of terminal clades Baccatum (C. baccatum, C. praetermissum, C. chacoense) and Annuum (C. annuum, C. chinense, C. frutescens) -highly supported (0.93)- at the NTS ML tree coincides with the low to high support found in previous analysis (Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
). Capsicum tovarii is a pungent chili pepper also used as a spice (Eshbaugh et al. 1983Eshbaugh WH, Smith PG & Nickrent DL (1983) Capsicum tovarii (Solanaceae), a new species of pepper from Peru. Brittonia 35: 55-60. DOI: 10.2307/2806051
https://doi.org/10.2307/2806051...
) and is particular in that solely produce fertile seeds when cross hybridizes with members of the Baccatum clade than to the Annuum clade -in spite of the above stated- or even C. pubescens and C. eximium (Tong & Bosland 1999Tong N & Bosland PW (1999) Capsicum tovarii, a new member of the C. baccatum complex. Euphytica 109: 71-77. DOI: 10.1023/A:1003421217077
https://doi.org/10.1023/A:1003421217077...
).
ML phylogenetic tree of the 5S NTS sequences of Capsicum and comparable related taxa -Lycianthes, Solanum, Datura, Atropa-. Phylogenetic relationships were inferred via FastTree 2.1.5 employing the NTS alignment matrix of Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Fig. 12, the GTR+G model and 1,000 resamples. Selected support values are shown side to main branches and those below 50% are condensed. For a detailed support values depiction see Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Fig. 13. Note the grouping of the NTS sequences at expected clades in Capsicum.
The germplasm of major consumed domesticated chili peppers -C. pubescens, C. baccatum var. umbilicatum, C. baccatum var. pendulum, C. frutescens, C. chinense, C. annuum var. annuum- and their related wild varieties -C. baccatum var. baccatum, C. annuum var. glabriusculum- was further investigated through a multiple alignment of 43 NTS sequences (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 1) and a subsequent bootstrap phylogenetic network (Fig. 8a). As a central result, different species could be discriminated according to sequence traits such as SNPs -fixed and majority- and indels. In agreement, NTS sequences at the network also clustered into well recognized species categories, including previous data (Park et al. 2000Park YK, Park KC, Park CH & Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Molecules and Cells 10: 18-24.). What is more, ancestral and derived traits at the middle SRII on paralogs of C. pubescens did not disrupt their clustering. In addition, NTS sequences of C. annuum fall into two groups, 1) wild variety paralogs and 2) all domesticated plus wild variety paralogs, as evidence of selection processes during domestication (Fig. 8a). With respect to C. baccatum, both sequence traits and network analysis proved useful to discriminate among varieties umbilicatum and pendulum whereas -as expected for selection- wild variety paralogs separated in those groups. Hence, NTS based variety discrimination in chili peppers constitutes a promising tool, similar to tomato (Sun et al. 2014bSun YL, Kang HM, Kim YS, Baek JP, Zheng SL, Xiang J-J & Hong S-K (2014b) Tomato (Solanum lycopersicum) variety discrimination and hybridization analysis based on the 5S rRNA region. Biotechnology and Biotechnological Equipment 28: 431-437. DOI: 10.1080/13102818.2014.928499
https://doi.org/10.1080/13102818.2014.92...
). Further, worth noting here that the discrimination value of the NTS extends to overall major consumed chili peppers, including the wild C. chacoense and C. eximium.
a-b. Bootstrap phylogenetic network of the 5S NTS sequences of domesticated and their related wild varieties of Capsicum, and of Atlantic Forest clade members- a. of domesticated and their related wild varieties of Capsicum; b. of Atlantic Forest clade members. Networks were constructed via SplitsTree4 employing the NTS alignment matrices of Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Fig. 1 (for a.) and Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Fig. 2 (for b.), the Neighbornet algorithm, the uncorrected P distance and 100 boostrap replicates. pub and pubAF217954 = C. pubescens; bac = C. baccatum var. baccatum; umb and bacAF217951 = C. baccatum var. umbilicatum; pen = C. baccatum var. pendulum; fru and fruAF217952 = C. frutescens; chi and chiAF217953 = C. chinense; gla = C. annuum var. glabriusculum; ann and annAF217950 = C. annuum var. annuum; per = C. pereirae; vil = C. villosum; fri = C. friburgense; sch = C. schottianum; rec = C. recurvatum; cam = C. campylopodium; hun = C. hunzikerianum. Scale bar indicates the scale of the network branches.
Atlantic Forest clade members are distinct in having small pungent yellowish green fruits, in addition to 2n = 26 chromosomes and highly variable karyotypes useless to solve species relationships (Pozzobon et al. 2006Pozzobon MT, Schifino-Wittmann MT & Bianchetti LDB (2006) Chromosome numbers in wild and semidomesticated Brazilian Capsicum L. (Solanaceae) species: do x=12 and x=13 represent two evolutionary lines? Botanical Journal of the Linnean Society 151: 259-269. DOI: 10.1111/j.1095-8339.2006.00503.x
https://doi.org/10.1111/j.1095-8339.2006...
; Moscone et al. 2007Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE & Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Horticulturae 745: 137-169. DOI: 10.17660/ActaHortic.2007.745.5
https://doi.org/10.17660/ActaHortic.2007...
). Analysis on taxonomy and partial phylogeny of its species were also reported (Buso et al. 2002Buso GSC, Amaral ZPS, Bianchetti LDB & Ferreira ME (2002) Análise de seqüências de DNA cloroplástico de espécies do gênero Capsicum. Boletim de Pesquisa e Desenvolvimiento. Vol. 37. Embrapa Recursos Genéticos e Biotecnologia, Brasilia. 15p., 2003Buso GSC, Amaral ZPS, Bianchetti LDB, Machado FRB & Ferreira ME (2003) Genetic variability and phylogenetic analysis of brazilian species of Capsicum. Capsicum and Eggplant Newsletters 22: 13-16.; Barboza & Bianchetti 2005Barboza GE & Bianchetti LDB (2005) Three new species of Capsicum (Solanaceae) and a key to the wild species from Brazil. Systematic Botany 30: 863-871. DOI: 10.1600/036364405775097905
https://doi.org/10.1600/0363644057750979...
). The most complete analysis also found difficulties to unambiguously solve species relationships in this clade, arguing that it is in an scenario of apparent phase of rapid speciation (Carrizo García et al. 2016Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
).
In this work, the Atlantic Forest clade revealed as well supported, however both global NTS phylogenetic approaches found the same question to solve species relationships (Figs. 6; 7). In an effort to clarify this issue, we performed an additional multiple alignment of 39 NTS sequences of this clade, which was reference compared to Lycianthes in order to identify synapomorphies (Grabiele et al. 2020Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1...
, Suppl. Fig. 2) and further built a bootstrap phylogenetic network for the seven species (Fig. 8b). Twelve synapomorphic traits were found and sequences clustered into an ancestral state group -C. pereirae, C. villosum, C. friburgense- and a derived state group -C. recurvatum, C. campylopodium, C. hunzikerianum- while C. schottianum paralogs fall into both groups. Despite the coexistence of sequences combining ancestral and derived traits in the rDNA array, synapomorphies indicate that C. schottianum is closely related to C. recurvatum, C. campylopodium and C. hunzikerianum than to others.
At this point, the NTS revealed useful to circumscribe the Atlantic Forest clade into two phylogenetically relevant groups of species. In this sense, our contribution is major since Atlantic Forest clade species could be attractive as a source of variability, however studies regarding potentially valuable genes, i.e. disease resistance and abiotic stress tolerance, reproductive biology, and fruits and their uses, are still lacking for this group. Further, cross hybridization attempts between an Atlantic Forest clade member (as C. buforum; probably misinterpreted n = 12) and chili peppers of distinct clades were reported as unsuccessful (Tong & Bosland 2003Tong N & Bosland PW (2003) Observations on interspecific compatibility and meiotic chromosome behavior of Capsicum buforum and C. lanceolatum. Genetic Resources and Crop Evolution 50: 193-199. DOI: 10.1023/A:1022986615694
https://doi.org/10.1023/A:1022986615694...
), analysis that should be extended to the whole group. In this scenario, gene transference between the Atlantic Forest clade members and the main consumed chili peppers based on x = 12 could be overcome biotechnologically, i.e. by the embryo rescue technique (Tripodi & Kumar 2019Tripodi P & Kumar S (2019) The Capsicum crop: an introduction. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 1-8. DOI: 10.1007/978-3-319-97217-6).
Combined data on genetic variability and phylogeny of the NTS in Capsicum
Sequence information of each structural region (SRI-III) was further integrated onto the NTS ML phylogenetic tree. In this sense, combined data revealed main evolutionary features at the NTS of Capsicum, and this molecular variability that includes insertions, deletions and substitutions, proved useful to circumscribe species or group of species (Fig. 9).
Main evolutionary features at the 5S NTS of Capsicum onto the ML phylogenetic tree. a = 3’ end SRI: novel CT block; middle SRII: novel CGGG motif. b = Middle SRII: major insertion (13 nt). c = 5’ end SRI: major deletion (21 nt) not affecting polyT stretch; CT block CTCTCC-type. d = SRII: almost entire deletion (C. coccineum); CT block MCTCTC-type. e = Middle SRII: partial deletion; CT-block CGTTCTGT-type. f = Near 5’ end SRII: TT to two purines. g = SRII: major block deletions, CGGG to CGGGG and two purines insertion at 3’ end; SRIII: GAT to GAC at 3’ end; CT block CCTCTT-type. h = Middle SRI: AATTT to GATTT, CT insertion, GCG to GTG; Near 5’ end SRII: C to G. i = SRI: deletion of TGTCG and insertion of a large polyT stretch (16 nt group parsimony) upstream ancestral polyT stretch (CCTTTTT). j = SRI: major insertion (123 nt) of a 5S related stretch near 3’ end. k = Middle SRIII: AGA to GGA. l = SRI: polyT stretch insertion of 16 to 21 nt (group parsimony). m = SRI: polyT stretch insertion of 16 to 12 nt (group parsimony); T to A at 3’ end of polyT stretch. n = SRI: insertion (7 nt) of repetitive nature at polyT stretch (C. chacoense). o = Near 5’ end SRII: major purine-rich block deletion (12 nt; C. praetermissum).
To further integrate the NTS ML phylogenetic tree with another component of the molecular variability, nucleotide substitution rates at the NTS of Capsicum and comparable related taxa were estimated in view of phylogenetically relevant groups and their splits time (Sarkinen et al. 2013Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
https://doi.org/10.1186/1471-2148-13-214...
) (Fig. 10). This way, substitution rate at the split of the non pungent Andean clade and the rest of Capsicum (3.33e-08) occurring 9.8 MYA is significantly lower -near half- than post split rates averaging 6.00e-08 (4.65e-08 to 8.45e-08). In this sense, there appear to emerge a correlation between accelerated substitution rates at the NTS and the recent diversification of the genus (5.38-3.44 MYA). This diversification, gave birth to the Atlantic Forest, Flexuosum, Bolivian, Pubescens and Purple Corolla clades, so as to the common ancestor of Tovarii, Baccatum and Annuum clades (TBA). Then, substitution rates remain constant during the divergence of TBA group, to which indels also contributed. Meanwhile, an almost invariable substitution rate through a period of 3.66 million years (5.09e-08 to 4.65e-08) and a nucleotide diversity value (π = 0.080) that is the lowest among analyzed groups support the proposed scenario of current rapid speciation at the Atlantic Forest clade. Contrary, substitution rate at the NTS increased significantly (1.04e-07) at the recent diversification of the Andean clade 3.09 MYA, and also at the Annuum clade (8.45e-08) during the course that led to the three major cultivated species of Capsicum. In a broad sense, the clockwise scenario of expansion and diversification of chili peppers hypothesized by Carrizo García et al. (2016)Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
https://doi.org/10.1093/aob/mcw079...
is supported here through combined NTS structural and evolutionary features, which reinforces the value of the 5S NTS as a marker with phylogenetic relevance.
Nucleotide substitution rates at the 5S NTS of Capsicum and comparable related taxa -Lycianthes, Solanum, Datura, Atropa-. Nucleotide diversity (π) of phylogenetically relevant groups was estimated in MEGA 7.0.26 employing the NTS alignment matrix of Grabiele et al. (2020)Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
http://dx.doi.org/10.17632/xcrmb7m7y8.1... , Suppl. Fig. 12, the K2+G model and 100 bootstrap replicates. Evolutionary divergence times in Solanaceae are considered (Sarkinen et al. 2013Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
https://doi.org/10.1186/1471-2148-13-214... ). SR = substitution rate; s.e. = standard error; MYA = million years ago; ROT = rest of taxa; ROC = rest of clades; CA = common ancestor. Vertical bars correspond to s.e. of SR.
Our analysis is original since includes all domesticated species of Capsicum prevailing in germplasm collections and breeding programs, together with a large group of wild chili peppers that demanded further genetic characterization. The 5S rDNA NTS demonstrated as a reliable and efficient genetic marker to characterize variability among chili peppers. NTS nucleotide sequences and their structural evolutionary traits such as indels and single nucleotide polymorphisms (SNP) alone proved useful to circumscribe taxa. In this sense, the attained comprehensive multiple alignments, now available to the interested community, serve as a taxa identification toolkit for chili peppers: it depends only in traditional and cost effective PCR amplification and Sanger sequencing, providing a set of valuable markers to germplasm managers and breeders. At the same time, the NTS is a valuable tool to reconstruct evolutionary relationships in Capsicum to a broad taxonomical range, via classical phylogenetic tree and alternative phylogenetic network approaches. The evolutionary scenario based on the NTS confirm monophyly, independent origin of both x = 13 lineages, major clades subdivision and diversification in Capsicum. Horticulture of chili peppers can benefit of this double purpose genetic marker, as well also the germplasm characterization and evolutionary studies within Solanaceae.
Acknowledgements
This study was funded by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT-Argentina), UNaM PICT 2014-3328 Préstamo BID Nº AR-L 1181. We would like to thank Dr. Gloria E. Barboza (IMBIV-UNC-CONICET), for her expertise in the identification of the plant material. This contribution is dedicated to the memory of Dr. Eduardo A. Moscone, pioneer in chili peppers molecular cytogenetics and mentor of this study.
References
- Aguilera PM, Debat HJ, Scaldaferro MA, Martí DA & Grabiele M (2016) FISH-mapping of the 5S rDNA locus in chili peppers (Capsicum-Solanaceae). Anais da Academia Brasileira de Ciências 88: 117-125. DOI: 10.1590/0001-37652301620140616
» https://doi.org/10.1590/0001-37652301620140616 - Ahn Y-K, Manivannan A, Karna S, Jun T-H, Yang E-Y, Choi S, Kim J-H, Kim D-S & Lee E-S (2018) Whole genome resequencing of Capsicum baccatum and Capsicum annuum to discover single nucleotide polymorphism related to powdery mildew resistance. Scientific Reports 8: 5188. DOI: 10.1038/s41598-018-23279-5
» https://doi.org/10.1038/s41598-018-23279-5 - Allaby RG & Brown TA (2001) Network analysis provides insights into evolution of 5S rDNA arrays in Triticum and Aegilops Genetics 157: 1331-1341.
- Barboza GE & Bianchetti LDB (2005) Three new species of Capsicum (Solanaceae) and a key to the wild species from Brazil. Systematic Botany 30: 863-871. DOI: 10.1600/036364405775097905
» https://doi.org/10.1600/036364405775097905 - Barboza GE, Carrizo García C, Leiva González S, Scaldafferro M & Reyes X (2019) Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS One 14: e0209792. DOI: 10.1371/journal.pone.0209792
» https://doi.org/10.1371/journal.pone.0209792 - Barboza GE, de Bem Bianchetti L & Stehmann JR (2020) Capsicum carassense (Solanaceae), a new species from the Brazilian Atlantic Forest. PhytoKeys 140: 12-138. DOI: 10.3897/phytokeys. 140.47071
» https://doi.org/10.3897/phytokeys. 140.47071 - Barchenger DW, Naresh P & Kumar S (2019) Genetic resources of Capsicum In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 9-23. DOI: 10.1007/978-3-319-97217-6
- Besendorfer V, Krajacic-Sokol I, Jelenie S, Puizina J, Mlinarec J, Sviben T & Papes D (2005) Two classes of 5S rDNA unit arrays of the silver fir, Abies alba Mill.: structure, organization and evolution. Theoretical and Applied Genetics 110: 730-740. DOI: 10.1007/s00122-004-1899-y
» https://doi.org/10.1007/s00122-004-1899-y - Buso GSC, Amaral ZPS, Bianchetti LDB & Ferreira ME (2002) Análise de seqüências de DNA cloroplástico de espécies do gênero Capsicum Boletim de Pesquisa e Desenvolvimiento. Vol. 37. Embrapa Recursos Genéticos e Biotecnologia, Brasilia. 15p.
- Buso GSC, Amaral ZPS, Bianchetti LDB, Machado FRB & Ferreira ME (2003) Genetic variability and phylogenetic analysis of brazilian species of Capsicum Capsicum and Eggplant Newsletters 22: 13-16.
- Cardoso R, Ruas CF, Giacomin RM, Ruas PM, Ruas EA, Barbieri RL, Rodrigues R & Gonçalves LSA (2018) Genetic variability in Brazilian Capsicum baccatum germplasm collection assessed by morphological fruit traits and AFLP markers. PLoS ONE 13: e0196468. DOI: 10.1371/journal.pone.0196468
» https://doi.org/10.1371/journal.pone.0196468 - Carles M, Cheung MKL, Moganti S, Dong TTX, Tsim KW, Ip NY & Sucher NJ (2005) A DNA microarrey for the authentication of toxic traditional chinese medicinal plants. Planta Medica 71: 580-584. DOI: 10.1055/s-2005-864166
» https://doi.org/10.1055/s-2005-864166 - Carrizo García C (2011) Fruit characteristics, seed production and pollen tube growth in the wild chilli pepper Capsicum flexuosum Flora 206: 334-340. DOI: 10.1016/j.flora.2010.05.008
» https://doi.org/10.1016/j.flora.2010.05.008 - Carrizo García C, Barfuss MHJ, Sehr EM, Barboza GE, Samuel R, Moscone EA & Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118: 35-51. DOI: 10.1093/aob/mcw079
» https://doi.org/10.1093/aob/mcw079 - Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S & Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytologist 168: 241-252. DOI: 10.1111/j.1469-8137.2005.01480.x
» https://doi.org/10.1111/j.1469-8137.2005.01480.x - Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillie C, Espagnol M-E, Picard G & Tourmente S (2002) Analysis of the 5S RNA Pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature RNA. Genome Research 12: 132-144. DOI: 10.1101/gr.181301
» https://doi.org/10.1101/gr.181301 - Colonna V, DʼAgostino N, Garrison E, Albrechtsen A, Meisner J, Facchiano A, Cardi T & Tripodi P (2019) Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Scientific Reports 9: 10067. DOI: 10.1038/s41598-019-46136-5
» https://doi.org/10.1038/s41598-019-46136-5 - De-Silva DL, Mota LL, Chazot N, Mallarino R, Silva Brandão KL, Gómez Piñerez LM, Freitas AVL, Lamas G, Joron M, Mallet J, Giraldo CE, Uribe S, Sarkinen T, Knapp S, Jiggins CD, Willmott KR & Elias M (2017) North Andean origin and diversification of the largest ithomiine butterfly genus. Scientific Reports 7: 45966. DOI: 10.1038/srep45966
» https://doi.org/10.1038/srep45966 - Drouin G & Moniz de Sá M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Molecular Biology and Evolution 12: 481-493. DOI: 10.1093/oxfordjournals.molbev.a040223
» https://doi.org/10.1093/oxfordjournals.molbev.a040223 - Eshbaugh WH, Smith PG & Nickrent DL (1983) Capsicum tovarii (Solanaceae), a new species of pepper from Peru. Brittonia 35: 55-60. DOI: 10.2307/2806051
» https://doi.org/10.2307/2806051 - Frasch M, Wenzel W & Hess D (1989) The nucleotide sequences of nuclear 5S rRNA genes and spacer regions of Petunia hybrida Nucleic Acids Research 17: 2857. DOI: 10.1093/nar/17.7.2857
» https://doi.org/10.1093/nar/17.7.2857 - Fulnecek J, Lim KY, Leitch AR, Kovarik A & Matyasek R (2002) Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88: 19-25. DOI: 10.1038/sj.hdy.6800001
» https://doi.org/10.1038/sj.hdy.6800001 - Fulnecek J & Kovarik A (2007) Low abundant spacer 5S rRNA transcripts are frequently polyadenylated in Nicotiana Molecular Genetics and Genomics 278: 565-573. DOI: 10.1007/s00438-007-0273-6
» https://doi.org/10.1007/s00438-007-0273-6 - Grabiele M, Debat HJ, Scaldaferro MA, Aguilera PM, Moscone EA, Seijo GJ & Ducasse DA (2018) Highly GC-rich heterochromatin in chili peppers (Capsicum-Solanaceae): a cytogenetic and molecular characterization. Scientia Horticulturae 238: 391-399. DOI: 10.1016/j.scienta.2018.04.060
» https://doi.org/10.1016/j.scienta.2018.04.060 - Grabiele M, Aguilera PM, Ducasse DA & Debat HJ (2020) Data for: Molecular characterization of the 5S rDNA non transcribed spacer and reconstruction of phylogenetic relationships in Capsicum. Mendeley Data. V1. Available at <http://dx.doi.org/10.17632/xcrmb7m7y8.1>. Access on 10 Sep. 2020. DOI: 10.17632/xcrmb7m7y8.1
» http://dx.doi.org/10.17632/xcrmb7m7y8.1 - Hori H, Lim B-L & Osawa S (1985) Evolution of green plants as deduced from 5S rRNA sequences. Proceedings of the National Academy of Sciences of the United States of America 82: 820-823. DOI: 10.1073/pnas.82.3.820
» https://doi.org/10.1073/pnas.82.3.820 - Hori H & Osawa S (1987) Origin and evolution of organisms as deduced from 5S ribosomal RNA sequences. Molecular Biology and Evolution 4: 445-472. DOI: 10.1093/oxfordjournals.molbev.a040455
» https://doi.org/10.1093/oxfordjournals.molbev.a040455 - Huson DH & Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254-267. DOI: 10.1093/molbev/msj030
» https://doi.org/10.1093/molbev/msj030 - Ibarra-Torres P, Valadez-Moctezuma E, Pérez-Grajales M, Rodríguez-Campos J & Jaramillo-Flores ME (2015) Inter- and intraspecific differentiation of Capsicum annuum and Capsicum pubescens using ISSR and SSR markers. Scientia Horticulturae 181: 137-146. DOI: 10.1016/j.scienta.2014.10.054
» https://doi.org/10.1016/j.scienta.2014.10.054 - Ibiza VP, Blanca J, Cañizares J & Nuez F (2012) Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genetic Resources and Crop Evolution 59: 1077-1088. DOI: 10.1007/s10722-011-9744-z
» https://doi.org/10.1007/s10722-011-9744-z - Jarret RL, Barboza GE, Batista FRC, Berke T, Chou Y-Y, Hulse-Kemp A, Ochoa-Alejo N, Tripodi P, Veres A, Carrizo Garcia C, Csillery G, Huang Y-K, Kiss E, Kovacs Z, Kondrak M, Arce-Rodriguez ML, Scaldaferro MA & Szoke A (2019) Capsicum - an abbreviated compendium. Journal of the American Society for Horticultural Science 144: 3-22. DOI: 10.21273/JASHS04446-18
» https://doi.org/10.21273/JASHS04446-18 - Kazutaka K & Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772-780. DOI: 10.1093/molbev/mst010
» https://doi.org/10.1093/molbev/mst010 - Kellogg EA & Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140: 325-343.
- Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, Seo E, Choi J, Cheong K, Kim K-T, Jung K, Lee G-W, Oh S-K, Bae C, Kim S-B, Lee H-Y, Kim S-Y, Kim M-S, Kang B-C, Jo YD, Yang H-B, Jeong H-J, Kang W-H, Kwon J-K, Shin C, Lim JY, Park JH, Huh JH, Kim J-S, Kim B-D, Cohen O, Paran I, Suh MC, Lee SB, Kim Y-K, Shin Y, Noh S-J, Park J, Seo YS, Kwon S-Y, Kim HA, Park JM, Kim H-J, Choi S-B, Bosland PW, Reeves G, Jo S-H, Lee B-W, Cho H-T, Choi H-S, Lee M-S, Yu Y, Choi YD, Park B-S, van Deynze A, Ashrafi H, Hill T, Kim WT, Pai H-S, Ahn HK, Yeam I, Giovannoni JJ, Rose JKC, Sørensen I, Lee S-J, Kim RW, Choi I-Y, Choi B-S, Lim J-S, Lee Y-H & Choi D (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics 46: 270-278. DOI: 10.1038/ng.2877
» https://doi.org/10.1038/ng.2877 - Kim S, Park J, Yeom S-I, Kim Y-M, Seo E, Kim K-T, Kim M-S, Lee JM, Cheong K, Shin H-S, Kim S-B, Han K, Lee J, Park M, Lee H-A, Lee H-Y, Lee Y, Oh S, Lee JH, Choi E, Choi E, Lee SE, Jeon J, Kim H, Choi G, Song H, Lee JK, Lee S-C, Kwon J-K, Lee H-Y, Koo N, Hong Y, Kim RW, Kang W-H, Huh JH, Kang B-C, Yang T-J, Lee Y-H, Bennetzen JL & Choi D (2017) New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biology 18: 210. DOI: 10.1186/s13059-017-1341-9
» https://doi.org/10.1186/s13059-017-1341-9 - Kitamura S, Inoue M, Shikazono N & Tanaka A (2001) Relationships among Nicotiana species revealed by the 5S rDNA spacer sequence and fluorescence in situ hybridization. Theoretical and Applied Genetics 103: 678-686. DOI: 10.1007/s001220100643
» https://doi.org/10.1007/s001220100643 - Kumar S, Stecher G & Tamura K (2015) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870-1874. DOI: 10.1093/molbev/msw054
» https://doi.org/10.1093/molbev/msw054 - Kwon J-K & Kim B-D (2009) Localization of 5S and 25S rRNA genes on somatic and meiotic chromosomes in Capsicum species of chili pepper. Molecules and Cells 27: 205-209. DOI: 10.1007/s10059-009-0025-z
» https://doi.org/10.1007/s10059-009-0025-z - Matyasek R, Fulnecek J, Lim KY, Leitch AR & Kovarik A (2002) Evolution of 5S rDNA unit arrays in the plant genus Nicotiana (Solanaceae). Genome 45: 556-562. DOI: 10.1139/g02-017
» https://doi.org/10.1139/g02-017 - Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE & Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Horticulturae 745: 137-169. DOI: 10.17660/ActaHortic.2007.745.5
» https://doi.org/10.17660/ActaHortic.2007.745.5 - Onus AN & Pickersgill B (2004) Unilateral incompatibility in Capsicum (Solanaceae): occurrence and taxonomic distribution. Annals of Botany 94: 289-295. DOI: 10.1093/aob/mch139
» https://doi.org/10.1093/aob/mch139 - Park YK, Park KC, Park CH & Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Molecules and Cells 10: 18-24.
- Perina A, Seoane D, González-Tizón AM, Rodríguez-Fariña F & Martínez-Lage A (2011) Molecular organization and phylogenetic analysis of 5S rDNA in crustaceans of the genus Pollicipes reveal birth-and-death evolution and strong purifying selection. Evolutionary Biology 11: 304. DOI: 10.1186/1471-2148-11-304
» https://doi.org/10.1186/1471-2148-11-304 - Pozzobon MT, Schifino-Wittmann MT & Bianchetti LDB (2006) Chromosome numbers in wild and semidomesticated Brazilian Capsicum L. (Solanaceae) species: do x=12 and x=13 represent two evolutionary lines? Botanical Journal of the Linnean Society 151: 259-269. DOI: 10.1111/j.1095-8339.2006.00503.x
» https://doi.org/10.1111/j.1095-8339.2006.00503.x - Price MN, Dehal PS & Arkin AP (2010) FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. DOI: 10.1371/journal.pone.0009490
» https://doi.org/10.1371/journal.pone.0009490 - Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y, Yang Y, Wu Z, Mao L, Wu H, Ling-Hu C, Zhou H, Lin H, González-Morales S, Trejo-Saavedra DL, Tian H, Tang X, Zhao M, Huang Z, Zhou A, Yao X, Cui J, Li W, Chen Z, Feng Y, Niu Y, Bi S, Yang X, Li W, Cai H, Luo X, Montes-Hernández S, Leyva-González MA, Xiong Z, He X, Bai L, Tan S, Tang X, Liu D, Liu J, Zhang S, Chen M, Zhang L, Zhang L, Zhang Y, Liao W, Zhang Y, Wang M, Lv X, Wen B, Liu H, Luan H, Zhang Y, Yang S, Wang X, Xu J, Li X, Li S, Wang J, Palloix A, Bosland PW, Li Y, Krogh A, Rivera-Bustamante RF, Herrera-Estrella L, Yin Y, Yu J, Hu K & Zhang Z (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceedings of the National Academy of Sciences of the United States of America 111: 5135-5140. DOI: 10.1073/pnas.1400975111
» https://doi.org/10.1073/pnas.1400975111 - Reznikoff WS, Siegele DA, Cowing DW & Gross CA (1985) The regulation of transcription initiation in bacteria. Annual Review of Genetics 19: 355-387. DOI: 10.1146/annurev.ge.19.120185.002035
» https://doi.org/10.1146/annurev.ge.19.120185.002035 - Rivera A, Monteagudo AB, Igartua E, Taboada A, García-Ulloa A, Pomar F, Riveiro-Leira M & Silvar C (2016) Assessing genetic and phenotypic diversity in pepper (Capsicum annuum L.) landraces from North-West Spain. Scientia Horticulturae 203: 1-11. DOI: 10.1016/j.scienta.2016.03.006
» https://doi.org/10.1016/j.scienta.2016.03.006 - Rogers SO & Bendich AJ (1994) Extraction of total cellular DNA from plant, algae and fungi. In: Stanton BG & Schilperoort RA (eds.) A plant molecular biololgy manual. Kluwer Academic Publisher, Dordrecht. Pp. D1/1-8.
- Sambrook J & Russell DW (2001) Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 2100p.
- Sarkinen T, Bohs L, Olmstead RG & Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. DOI: 10.1186/1471-2148-13-214
» https://doi.org/10.1186/1471-2148-13-214 - Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, Holmes JB, Kim S, Kimchi A, Kitts PA, Lathrop S, Lu Z, Madden TL, Marchler-Bauer A, Phan L, Schneider VA, Schoch CL, Pruitt KD & Ostell J (2019) Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 47: D23-D28. DOI: 10.1093/nar/gky1069
» https://doi.org/10.1093/nar/gky1069 - Scaldaferro MA & Moscone EA (2019) Cytology and DNA content variation of Capsicum genomes. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 57-84. DOI: 10.1007/978-3-319-97217-6
- Stewart C, Mazourek M, Stellari GM, O’Connell M & Jahn M (2007) Genetic control of pungency in C. chinense via the Pun1 locus. Journal of Experimental Botany 58: 979-991. DOI: 10.1093/jxb/erl243
» https://doi.org/10.1093/jxb/erl243 - Sun Y-L, Choi I-L, Lee Y-B, Choi KY, Hong S-K & Kang H-M (2014a) Molecular diversity and phylogentic analysis of Capsicum annuum varieties using the nrDNA ITS region. Scientia Horticulturae 165: 336-343. DOI: 10.1016/j.scienta.2013.11.009
» https://doi.org/10.1016/j.scienta.2013.11.009 - Sun YL, Kang HM, Kim YS, Baek JP, Zheng SL, Xiang J-J & Hong S-K (2014b) Tomato (Solanum lycopersicum) variety discrimination and hybridization analysis based on the 5S rRNA region. Biotechnology and Biotechnological Equipment 28: 431-437. DOI: 10.1080/13102818.2014.928499
» https://doi.org/10.1080/13102818.2014.928499 - Sykorova E, Lim KY, Fajkus J & Leitch AR (2003) The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 112: 164-172. DOI: 10.1007/s00412-003-0256-2
» https://doi.org/10.1007/s00412-003-0256-2 - Szymanski M, Barciszewska MZ, Erdmann VA & Barciszewski J (2003) 5S rRNA: structure and interactions. Biochemical Journal 371: 641-651. DOI: 10.1042/BJ20020872
» https://doi.org/10.1042/BJ20020872 - Tong N & Bosland PW (1999) Capsicum tovarii, a new member of the C baccatum complex. Euphytica 109: 71-77. DOI: 10.1023/A:1003421217077
» https://doi.org/10.1023/A:1003421217077 - Tong N & Bosland PW (2003) Observations on interspecific compatibility and meiotic chromosome behavior of Capsicum buforum and C. lanceolatum Genetic Resources and Crop Evolution 50: 193-199. DOI: 10.1023/A:1022986615694
» https://doi.org/10.1023/A:1022986615694 - Tripodi P & Kumar S (2019) The Capsicum crop: an introduction. In: Ramchiary N & Kole C (eds.) The Capsicum genome. Springer, Cham. Pp. 1-8. DOI: 10.1007/978-3-319-97217-6
- Venkateswarlu K, Lee SW & Nazar RN (1991) Conserved upstream sequence elements in plant 5S ribosomal RNA-encoding genes. Gene 105: 249-253.
- Vizoso M, Vierna J, González-Tizón AM & Martínez-Lage A (2011) The 5S rDNA gene family in mollusks: characterization of transcriptional regulatory regions, prediction of secondary structures, and long-term evolution, with special attention to Mytilidae mussels. Journal of Heredity 102: 433-447. DOI: 10.1093/jhered/esr046
» https://doi.org/10.1093/jhered/esr046 - Volkov RA, Zanke C, Panchuk II & Hemleben V (2001) Molecular evolution of 5S rDNA of Solanum species (sect. Petota): application for molecular phylogeny and breeding. Theoretical and Applied Genetics 103: 1273-1282. DOI: 10.1007/s001220100670
» https://doi.org/10.1007/s001220100670 - Volkov RA, Panchuk II, Borisjuk NV, Hosiawa-Baranska M, Maluszynska J & Hemleben V (2017) Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna BMC Plant Biology 17: 21. DOI: 10.1186/s12870-017-0978-6
» https://doi.org/10.1186/s12870-017-0978-6 - Walsh BM & Hoot SB (2001) Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear waxy introns. International Journal of Plant Sciences 162: 1409-1418. DOI: 10.1086/323273
» https://doi.org/10.1086/323273
Edited by
Publication Dates
-
Publication in this collection
20 Sept 2021 -
Date of issue
2021
History
-
Received
07 Nov 2019 -
Accepted
09 July 2020