Abstracts
Bemisia tabaci (Genn.) is one of the most important pests in cultivated areas of vegetables and ornamental crops around the world. Based on the mitochondrial cytochrome oxidize I (mtCOI) sequence, there is evidence that B. tabaci should be considered a cryptic species complex of 11 groups containing 24 species. Two of the groups, Middle East-Asia Minor 1 and Mediterranean include biotypes B and Q, respectively. In this study we evaluated the mtCOI sequence of B. tabaci populations collected in sites of the state of São Paulo, Brazil. Using PCR-RFLP with Taq I, a typical biotype B profile was obtained for all specimens. Based on the comparison with mtCOI reference sequences we found four haplotypes all belonging to the Middle East-Asia Minor 1. They occurred in the hosts pepper (Capsicum annuum L.), tomato (Solanum lycopersicum L.), eggplant (Solanum melongena L.) and cucurbitaceae plants.
RFLP; cytochrome oxidase I gene; sequencing; whiteflies
Bemisia tabaci (Genn.) é considerada uma das mais importantes pragas em cultivos de hortaliças e ornamentais em todo o mundo. Baseado na análise da seqüência mitocondrial (citocromo oxidase I - mtCOI) foi proposto recentemente que B. tabaci deva ser considerado um complexo críptico de espécies, contendo 11 grupos e 24 espécies. Dois destes grupos: Middle East-Asia Minor e Mediterranean englobam os biótipos B e Q, respectivamente. Avaliou-se a sequência mtCOI de espécimes de B. tabaci coletados em regiões do estado de São Paulo, Brasil. Por PCR-RFLP utilizando-se a enzima Taq I, pôde-se observar somente o padrão típico de clivagem para o biótipo B. Comparando-se com sequências consenso, todas as moscas brancas foram classificadas no grupo Middle East-Asia Minor e puderam ser separadas em quatro haplótipos, indicando prevalência do biótipo B em áreas de pimentão (Capsicum annuum L.), tomate (Solanum lycopersicum L.), cucurbitáceas e berinjela (Solanum melongena L.) do Estado de São Paulo.
RFLP; gene do cytochromo oxidase I; seqüenciamento; moscas-brancas
NOTE
Only the B biotype of Bemisia tabaci is present on vegetables in São Paulo State, Brazil
Somente o biótipo B de Bemisia tabaci ocorre em hortaliças no Estado de São Paulo, Brasil
Kelly Cristina Gonçales RochaI; Julio Massaharu MarubayashiI; Jesús Navas-CastilloII; Valdir Atsushi YukiIII; Carlos Frederico WilckenI; Marcelo Agenor PavanI; Renate Krause-SakateI,* * Corresponding author < renatekrause@fca.unesp.br>
IUNESP - Depto. de Produção Vegetal, Setor Defesa Fitossanitária, C.P. 237 - 18603-970 - Botucatu, SP - Brasil
IIInstituto de Hortofruticultura Subtropical y Mediterránea "La Mayora"/UMA/CSIC, Estación Experimental "La Mayora" 29750 Algarrobo-Costa, Málaga, Spain
IIIInstituto Agronômico de Campinas, Centro de Fitossanidade - 13012-970 - Campinas, SP - Brasil
ABSTRACT
Bemisia tabaci (Genn.) is one of the most important pests in cultivated areas of vegetables and ornamental crops around the world. Based on the mitochondrial cytochrome oxidize I (mtCOI) sequence, there is evidence that B. tabaci should be considered a cryptic species complex of 11 groups containing 24 species. Two of the groups, Middle East-Asia Minor 1 and Mediterranean include biotypes B and Q, respectively. In this study we evaluated the mtCOI sequence of B. tabaci populations collected in sites of the state of São Paulo, Brazil. Using PCR-RFLP with Taq I, a typical biotype B profile was obtained for all specimens. Based on the comparison with mtCOI reference sequences we found four haplotypes all belonging to the Middle East-Asia Minor 1. They occurred in the hosts pepper (Capsicum annuum L.), tomato (Solanum lycopersicum L.), eggplant (Solanum melongena L.) and cucurbitaceae plants.
Key words: RFLP, cytochrome oxidase I gene, sequencing, whiteflies
RESUMO
Bemisia tabaci (Genn.) é considerada uma das mais importantes pragas em cultivos de hortaliças e ornamentais em todo o mundo. Baseado na análise da seqüência mitocondrial (citocromo oxidase I - mtCOI) foi proposto recentemente que B. tabaci deva ser considerado um complexo críptico de espécies, contendo 11 grupos e 24 espécies. Dois destes grupos: Middle East-Asia Minor e Mediterranean englobam os biótipos B e Q, respectivamente. Avaliou-se a sequência mtCOI de espécimes de B. tabaci coletados em regiões do estado de São Paulo, Brasil. Por PCR-RFLP utilizando-se a enzima Taq I, pôde-se observar somente o padrão típico de clivagem para o biótipo B. Comparando-se com sequências consenso, todas as moscas brancas foram classificadas no grupo Middle East-Asia Minor e puderam ser separadas em quatro haplótipos, indicando prevalência do biótipo B em áreas de pimentão (Capsicum annuum L.), tomate (Solanum lycopersicum L.), cucurbitáceas e berinjela (Solanum melongena L.) do Estado de São Paulo.
Palavras-chave: RFLP, gene do cytochromo oxidase I, seqüenciamento, moscas-brancas
Introduction
The whitefly Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae) is one of the most important pest of vegetables and ornamental crops in the world and is a vector of more than 100 plant viruses (Jones et al., 2003). Most of these viruses belong to the genus Begomovirus, one of the most important emerging groups of plant viruses (Brown, 2000). B. tabaci has a high intraspecific biological and genetic variability (De Barro et al., 2000). No morphological character, neither juvenile nor adult can distinguish different B. tabaci (Rosell et al., 1997; Calvert et al., 2001). Dinsdale et al. (2010) have recently proposed that B. tabaci should be considered as a cryptic species complex comprising 11 groups containing 24 species. The more reproducible and informative method available to determine the genetic affiliation of a B. tabaci population is sequencing the mitochondrial cytochrome oxidase I (mtCOI) gene (De Barro et al., 2005) and use consensus sequences to assign group affiliation (Dinsdale et al., 2010).
The Middle East-Asia Minor 1 group contains the B and B2 biotypes and the Mediterranean the Q, J, L, SubSaharan Africa Silverleaf biotypes (Dinsdale et al., 2010). The authors provide strong evidence that B and Q are different species not biotypes, but in this paper we will still consider them as different biotypes. The biotype B was introduced in Brazil in the beginning of the 1990's (Lourenção and Nagai, 1994) and in the last few years has spread to several states causing losses in many crops (Lima et al., 2002). Beside these biotype another were detected under Brazilian condition: biotype A or subsequently referred to as biotype BR by (Lima et al. 2000). The Q biotype has not yet been reported from Brazil (Rabello et al., 2008) and represents a threat due to its resistance to pyriproxyfen (Horowitz et al., 2003), buprofezin and reduced susceptibility to the neonicotinoid insecticides imidacloprid, acetamiprid and thimethoxam (Hodges, 2009), currently used to control B. tabaci.
The association of techniques like PCR-RFLP has also been used to successfully distinguish biotypes populations of B. tabaci (Bosco et al., 2006; Sartor et al., 2008). To determine whether the Q biotype is present in areas where tomato (Solanum lycopersicum L.) and pepper (Capsicum annuum L.) are grown in the state of São Paulo, B. tabaci samples were collected from these plants and the mtCOI gene was analyzed by PCR-RFLP and sequencing.
Material and Methods
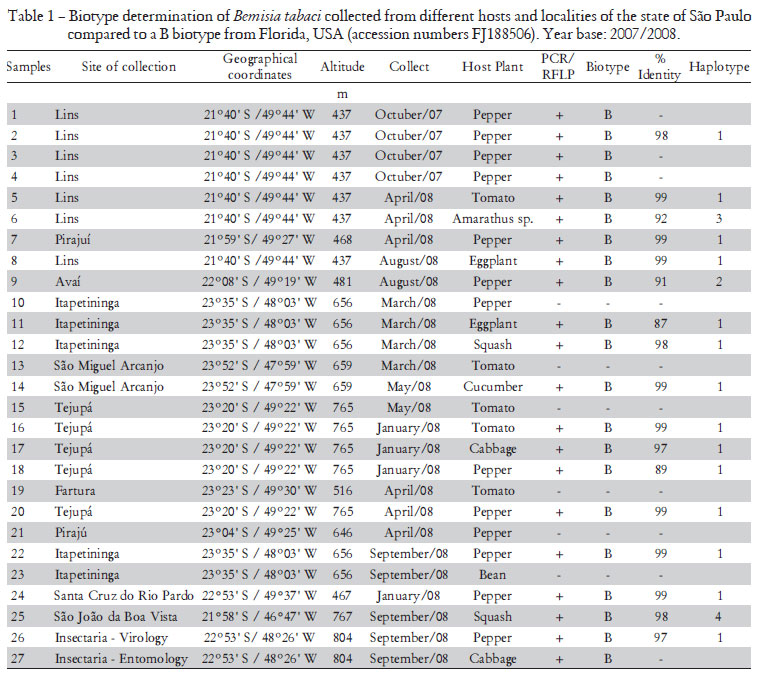
Adults of B. tabaci were collected from 27 localities in the state of São Paulo, Brazil, during 2007/2008, using a hand-held aspirator and preserved immediately in 95% ethanol at -20ºC until processing. Samples were collected mostly from pepper and tomato crops but also from other vegetables (Table 1). Colonies of B biotype from Brazil and the Q biotype from Spain were used as controls in the analysis.
Each individual sample was macerate directly in sterile Whatman 3 MM paper (2 mm2 pieces) which was placed on a 0.2 mL PCR tube. Part of the mtCOI gene was amplified with primers C1-J-2195-FW (5'-TTGATTTTTTGGTCATCCAGAAGT-3') and C1-J-2195-RV (5'-TCCAATGCACTAATCTGCCATATTA-3') (Simon et al., 1994; Frohlich et al., 1999). Analysis of PCR were performed in a final volume of 50 µL (MgCl2 50 mmol L-1, dNTP 2.5 µM, C1-J-2195 and MA483-RV 10 µM) using 0.5 Unit of Taq polymerase (Bioline). The amplification was carried out in a PCR Express Thermal Cycler (Bio-Rad) with the following steps: 5 min. at 94ºC (one cycle), followed by 35 cycles of 30 s at 94ºC, 45 s at 45ºC and 1 min. at 72ºC and a final extension step of 10 min.at 72ºC. The presence of mtCOI amplicons was visualized by electrophoresis in 1% agarose gel stained with ethidium bromide.
Five microliters of each amplified product (880bp) were digested with one unit of TaqI at 65ºC for 2h in a final volume of 15 µL, according to Bosco et al. (2006). The digested DNA was visualized by electrophoresis in 2% agarose gel stained with ethidium bromide. Five individuals of each population from each region were analyzed.
The mtCOI gene of seventeen representative populations was sequenced. Sequences were assembled and analyzed using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and compared against the consensus sequences of Dinsdale et al. (2010). The alignment of the sequences was performed using the Clustal W program (http://www.ebi.ac.uk/clustaw/#) (Thompson et al., 1994), and the phylogenetic tree was constructed using the MEGA 4.0 program (Kumar et al., 2004), using the Neighbor-joining method with a "bootstrap" value of 2000.
Results and Discussion
Analysis of PCR with mtCOI primers provided amplicons of the expected size of 880bp (Figure 1-A); five samples did not amplify with the mtCOI primers. These whiteflies were morphologically indistinguishable from those collected in other localities. These samples also failed to amplify when primed with the ribosomal ITS primers 5'-GTTTCCGTAGGTGAACCTGC-3' (Brust et al., 1998) and 5'-TCCGCGAGCCGAGTGATCC-3' (De Barro et al., 2000); it is therefore likely that the DNA was degraded. The digestion of the PCR product with TaqI showed the expected restriction patterns of the B biotype (Figure 1-B). The PCR-RFLP method used in this work easily distinguishes the B and Q biotypes (Bosco et al., 2006). The controls used in these experiments, a B biotype colony from Brazil, and a Q biotype colony from Spain also showed the expected restriction patterns.
The sequence of the mtCOI gene of some specimens representing the studied populations was determined. In the analysis by CLUSTAL W all the sequences had very high nucleotide (98% to 99%) identity with the mtCOI of the previously characterized B biotype presented in Argentina (accession numbers AF340215) and they could be assigned to one of four haplotypes (Table 1, Figure 2). A phylogenetic tree (Figure 2) was constructed using the mtCOI gene sequences obtained and consensus sequences for the different groups proposed by Dinsdale et al. (2010). All samples analyzed were classified in the Middle East-Asia Minor 1 genetic group that comprises specimens previously classified as biotype B (Figure 2). These results indicate that the Q biotype has not be established in Brazil. In addition, the high infestations of B. tabaci reported by Scarpellini et al. (2002) and Yuki (2001) in economically important annual crops and several vegetable crops in producing regions of the São Paulo state are probably of the B biotype. The BR biotype (Lima et al., 2000) was not found in this work.
Accurate identification of insects that are pests and/ or virus vectors is a prerequisite for their effective management to reduce crop damage (Brown, 2000). Analysis of the mitochondrial cytochrome oxidase I (mtCOI) gene of worldwide collections of the whitefly have greatly improved the understanding of the genetic diversity of the B. tabaci species complex (Bosco et al., 2006; Dinsdale et al., 2010). B. tabaci B biotype was the only biotype found in the main tomato and pepper cultivation areas of the state of São Paulo; this biotype also infested other hosts, such as cucumber (Cucumis sativus L.), squash (Cucurbita pepo L.), cabbage (Brassica oleracea L. var. acephala), eggplant (Solanum melongena L.) and Amaranthus spp. in this area. The knowledge gained from mtCO1 is an important tool that can help to prevent the introduction of exotic B. tabaci biotypes such as Q into Brazil, the invasion of alien species can cause serious economic, environmental and social impact on agricultural crops.
Received June 18, 2009
Accepted September 22, 2010
- Bosco, D.; Loria, A.; Sartor, C.; Cenis, J.L. 2006. PCR-RFLP Identification of Bemisia tabaci biotypes in the Mediterranean Basin. Phytoparasitica 34: 243-251.
- Brown, J.K. 2000. Molecular markers for the identification and global tracking of whitefly vector-begomovirus complexes. Virus Research 71: 233-260.
- Brust, R.A.; Ballard, J.W.O.; Driver, F.; Hartley, D.M.; Galway, N.J.; Curran, J. 1998. Molecular systematics, morphological analysis, and hybrid crossing identify a third taxon, Aedes (Halaedes) wardandensis sp. nov., of the Aedes (Halaedes) australis species-group (Diptera: Culicidae). Canadian Journal Zoology 76: 1236-1246.
- Calvert, L.A.; Cuervo, M.; Arroyave, J.A.; Constantino, L.M.; Bellotti, A.; Frohlich, D. 2001. Morphological and mitochondrial DNA marker analyses of whiteflies (Homoptera: Aleyrodidae) colonizing cassava and beans in Colombia. Annals of the Entomological Society of America 94: 512-519.
- De Barro, P.J.; Drive, F.; Trueman, J.W.; Curran, J. 2000. Phylogenetic relationships of world populations of Bemisia tabaci (Gennadius) using ribosomal ITS1. Molecular Phylogenetic Evolution 16: 29-36.
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) Mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Annals of the Entomological Society of America 103: 196-208.
- Frohlich, D. R., Torres-Jerez, I., Bedford, I. D., Markham, P. G. & Brown, J. K., 1999. A phylogeographical analysis of the Bemisia tabaci species complex based on mitocondrial DNA markers. Molecular Ecolology 8: 1683-1691.
- Horowitz, A.; Gormon, K.; Ross, G.; Denholm, I. 2003. Inheritance of pyriproxyfen resistance in the whitefly, Bemisia tabaci (Q biotype). Archives of Insect Biochemistry and Physiology 54: 177-186.
- Jones, D.R. 2003. Plant viruses transmitted by whiteflies. European Journal of Plant Pathology 109: 195-219.
- Kumar, S.; Tamura, K.; Nei, M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5: 150-163.
- Lima, L.H.C.; Navia, D.; Inglis, P.W.; Oliveira, M.R.V.; Oliveira, M.R.V. 2000. Survey of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) biotypes in Brazil using RAPD markers. Genetics and Molecular Biology 23: 781-785.
- Lima, L.H.C.; Campos, L.; Moretzsohn, M.C.; Navia, D.; Oliveira, M.R.V. 2002. Genetic diversity of Bemisia tabaci (Genn.) populations in Brazil revealed by RAPD markers. Genetics and Molecular Biology 25: 217-223.
- Lourenção, A.L.; Nagai, H. 1994. Outbreaks of Bemisia tabaci in the São Paulo State, Brazil. Bragantia 53: 53-59. (in Portuguese with abstract in English).
- Rabello, A.R.; Queiroz, P.R.; Simões, K.C.C.; Hiragi, C.O.; Lima, L.H.C.; Oliveira, M.R.V.; Mehta, A. 2008. Diversity analysis of Bemisia tabaci biotypes: RAPD, PCR-RFLP and sequencing of the ITS1 rDNA region. Genetics and Molecular Biology 31: 585-590.
- Rosell, R.C.; Bedford, I.D.; Frohlich, D.R.; Gill, R.J.; Brown, J.K. 1997. Analysis of morphological variation in distinct populations of Bemisia tabaci (Homoptera: Aleyrodidae). Annals of the Entomological Society of America 90: 575-589
- Sartor, C.; Demichelis, S.; Cenis, J.L.; Coulibaly, A.K.; Bosco, D. 2008. Genetic variability of Bemisia tabaci in the Mediterranean and Sahel Regions. Bulletin of Insectology 61: 161-162.
- Scarpellini, J.R.; Ramiro, Z.A.; Lara, R.I.R.; Santos, J.C.C. 2002. Chemical control of whitefly Bemisia tabaci (Gennadius, 1889) B biotype (Hemiptera: Aleyrodidae) on common beans. Arquives of Institute Biological 69: 23-27. (in Portuguese with abstract in English).
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compliation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America 87: 651-701.
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673-4680.
- Yuki, V.A. 2001. Whitefly: history of outbreaks and control measures as pest and vector of viruses. O Agronômico 53: 23-25. (in Portuguese).
Publication Dates
-
Publication in this collection
13 Jan 2011 -
Date of issue
Feb 2011
History
-
Accepted
22 Sept 2010 -
Received
18 June 2009