Abstract
Bemisia tabaci (Genn.) represents a complex of cryptic species that causes losses of many valuable crops. Even though there are differences in their ability to colonize hosts, transmit phytovirus and develop resistance to insecticides, the genetic differentiation of the insect populations is important for the adoption of control measures. Therefore, the genetic diversity of B. tabaci populations in economically important crops in Brazilian locations was characterized through a microsatellite analysis. Eight microsatellite markers were used for the analysis of eight populations, three in Solanum tuberosum (States of São Paulo, Minas Gerais and Bahia), two in Glycine max (States of São Paulo and Mato Grosso), one in Phaseolus vulgaris and Brassica oleracea var. acephala (States of São Paulo and Distrito Federal, respectively) and a cabbage population from Florida (USA). The number of alleles varied between two and 13 and the average value of F ST was 0.13. The population occurring in beans was genetically different, suggesting that the excessive use of insecticide or the host itself may have caused the modification of its allele frequency. The American population presented a large diversity and small differentiation compared to the Brazilian populations, especially from the Southeast, supporting the hypothesis that the B biotype was probably introduced into Brazil by the trade of plant material between the USA and the State of São Paulo. The genetic diversity found within and among the populations is geographically structured, and the insects from the central region of Brazil had superior genetic divergence when compared to the others Brazilian locations.
SSR; biotype B; population genetics; silverleaf whitefly
ENTOMOLGY
Structure of genetic diversity of Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae) populations in Brazilian crops and locations
Fernanda von Hertwig Mascarenhas FontesI; Carlos Augusto ColomboII, * * Corresponding author ccolombo@iac.sp.gov.br ; André Luiz LourençãoI
IIAC/Centro de Fitossanidade, C.P. 28 - 13020-970 - Campinas, SP - Brasil
IIIAC/Centro de Genética, C.P. 28 - 13020-970 - Campinas, SP - Brasil
ABSTRACT
Bemisia tabaci (Genn.) represents a complex of cryptic species that causes losses of many valuable crops. Even though there are differences in their ability to colonize hosts, transmit phytovirus and develop resistance to insecticides, the genetic differentiation of the insect populations is important for the adoption of control measures. Therefore, the genetic diversity of B. tabaci populations in economically important crops in Brazilian locations was characterized through a microsatellite analysis. Eight microsatellite markers were used for the analysis of eight populations, three in Solanum tuberosum (States of São Paulo, Minas Gerais and Bahia), two in Glycine max (States of São Paulo and Mato Grosso), one in Phaseolus vulgaris and Brassica oleracea var. acephala (States of São Paulo and Distrito Federal, respectively) and a cabbage population from Florida (USA). The number of alleles varied between two and 13 and the average value of FST was 0.13. The population occurring in beans was genetically different, suggesting that the excessive use of insecticide or the host itself may have caused the modification of its allele frequency. The American population presented a large diversity and small differentiation compared to the Brazilian populations, especially from the Southeast, supporting the hypothesis that the B biotype was probably introduced into Brazil by the trade of plant material between the USA and the State of São Paulo. The genetic diversity found within and among the populations is geographically structured, and the insects from the central region of Brazil had superior genetic divergence when compared to the others Brazilian locations.
Keywords: SSR, biotype B, population genetics, silverleaf whitefly
Introduction
Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae) belongs to a group of insects named whiteflies known throughout the world for their great capacity to cause serious damage to various crops (Brown et al., 1995). It is a polyphagous insect, and at least 600 species of plants are considered its hosts (Oliveira, 2001). Both nymphs and adults feed on plant sap, and can cause yellowing, mottling and leaf fall. The excretion of a substance known as honeydew facilitates fungus (sooty mold) to colonize the leaf surface, which may affect the development of the plant (Byrne and Bellows, 1991). Besides, B. tabaci is vector for more than 110 phytoviruses (Jones, 2003).
Since it became a pest in agriculture, this insect has been the target of genetic, morphological and behavioral studies. B. tabaci probably comprises a species complex whose morphological resemblance makes it difficult to differentiate them through conventional taxonomy (Brown et al., 1995). Recently, Dinsdale et al. (2010) analyzed data from 454 individuals by sequencing the mitochondrial cytocrome oxidase I gene (mtCOI) (DeBarro et al., 2005) and proposed that B. tabaci is a cryptic species complex comprising 11 groups that contain 24 species. De Barro et al. (2011) conducted crossing experiments that demonstrated reproductive incompatibility among some putative species (or biotypes). Despite this, as there is still no consensus for a new nomenclature to use, we will continue to adopt the term biotype whenever necessary.
In Brazil, infestation outbreaks of this pest in the beginning of the 1990's turned out to be related to the B biotype, that displaced the existing biotype A in the country (Lourenção and Nagai, 1994). Since then, through genetic studies with RAPD molecular markers (Lima et al., 2000; Lima et al., 2002; Fontes et al., 2010) and mitochondrial gene sequences (Rocha et al., 2011), no evidence was obtained concerning the presence of other biotypes in the country. Therefore, the level and structure of the genetic diversity for this insect in many crops of economic importance in Brazilian locations was characterized, aiming to understand the genetic relationship among them, through the use of microsatellite markers (SSR). This was the first study to use a population-based approach for B. tabaci in Brazil.
Materials and Methods
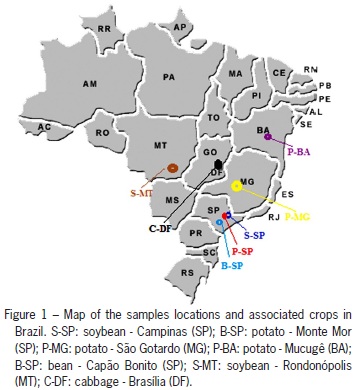
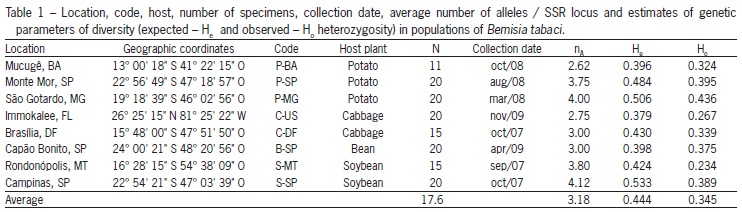
B. tabaci populations from seven locations in Brazil and one in the United States were collected, totaling 141 individuals (Table 1 and Figure 1). Immokalee, FL, US (26°25'15" N; 81°25'22" W) and Campinas, SP (22°54'21" S; 47°03'39" O) populations were previously characterized as biotype B. The insects used for this study were collected alive in the field, and were immediately stored in 92 % ethanol and kept refrigerated until used. The species is a haploi-diploid and based on this only females were used because they are diploid and can reveal more genetic information at a locus.
For obtaining total DNA, each adult female individual was macerated in 1.5 mL tubes containing 60 µL of lysis buffer (Tris-HCl 10 mM pH 8.0, EDTA 1 mM, Triton X-100 0.3 %, proteinase K 60 µg mL-1) and incubated at 65 ºC for 20 min. After that, the homogenate was incubated at 95 ºC for 7 min (to inactivate the proteinase K) and frozen at - 20 ºC (Aljanabi et al., 1998 modified by Lima et al., 2002). The concentration adopted was 3 ng µL-1 of the homogenate.
Simple sequence repeat (SSR)-anchored polymerase chain reaction amplification was adopted and eight microsatellite loci were considered in this study (Table 2). The PCR reactions were conducted in the final volume of 20 µL, containing from 10 to 15 ng of DNA; 0.4 µM from forward primer labeled with fluorochrome and reverse primer; 0.2 mM of each deoxyribonucleoside triphosphate (dNTP); 1 X PCR buffer (50 mM of KCl, 10 mM of Tris-HCl, pH 8.8); 1 U of Taq DNA polymerase and 1.5, 2 or 4 mM MgCl2. Primers sequences and differential PCR conditions are shown in Table 2. Amplifications of the loci followed the same cycling in a thermal cycler PTC 200 (MJ Research Inc.) programmed to start with 5 min at 94 °C for initial denaturation, followed by 30 cycles with 1 min denaturation at 94 °C, 1 min in the annealing temperature (Table 2), 1 min for extension at 72 ºC and 8 min at 72 °C for final extension. Amplification products were separated under denaturing conditions in 5 % polyacrylamide gels containing 8 M urea and 1 X TBE (Tris borate-EDTA) in an automated sequencer (ABI 377) and size of alleles obtained by the Genotyper® software version 2.0 (Applied Biosystems, Foster City, CA).
The allelic and genotypic frequencies at each locus were subjected to the Fisher's exact test with the proportions of the Hardy-Weinberg equilibrium, as defined by Weir (1996), using the program GENEPOP version 2.0 (Raymond and Rousset, 1995). Fisher's exact test was carried out by the Markov Chain method using 20 batches with 5000 permutations per batch. Genetic diversity and F statistics were estimated using a randomized design, according to Weir and Cockerham (1984). Allele frequencies, the number of alleles per locus (A), the heterozygosity observed (Ho) and expected (He) and Wright's F statistics (FIS, FIT and FST) were estimated using the Genetix version 4.05 program (Belkhir et al., 1996-2004). Bonferroni corrections were performed before proceeding to tests of significance of Fst values (Rice, 1989).
Structure software version 2.2 (Pritchard et al., 2000), based on Bayesian statistics´ was alternatively used to infer the number of groups (K). For this the value of K were tested ranging from one to ten runs. Simulations were done by 50,000 burn-ins and 500,000 Monte Carlo and Markov Chains (MCMC). To compare the most likely and proposed K definitions, the criteria adopted were proposed by Pritchard and Wen (2004) and Evanno et al. (2005). The structure of the variability was visualized by a dendrogram constructed from the genetic distance matrix of Nei (1978) and the UPGMA clustering criterion, using the NTSYS program (Rohlf, 1989) for analysis. The same program was used to calculate the significance of the correlation between genetic distance and geographical distance by the Mantel test (Mantel, 1967). For principal component analysis (PCA), the genetic distance of Nei (1978) was utilized and the calculations performed using Statistica software (Statsoft, 1999).
Results and Discussion
A total of 47 alleles were detected at all eight loci, analyzed ranging from two to 13 alleles (Table 1), similar to that obtained by Tsagkarakou and Roditakis (2003), who examined six populations of B. tabaci represented by 18 individuals each, with the same molecular marker and two loci in common with our work (BT4 and BTb-103).
The average He was superior to Ho for all populations (Table 1), which shows a deficit of heterozygotes for this species. Simón et al. (2007) explain heterozygote deficiency by the presence of null alleles, i.e., an allele that is present in a sample, but is not amplified because of some site mutation that inhibit amplification for the non binding of the primer in the DNA, and the result can be a false homozygote (Kwok et al., 1990). Another explanation is based on biological aspects of the insect, since it is a preferential colonizer of annual cycle plants, whose colonization is made primarily by few genotypes increasing the chances of inbreeding, thus promoting the heterozygote deficiency observed (De Barro, 2005).
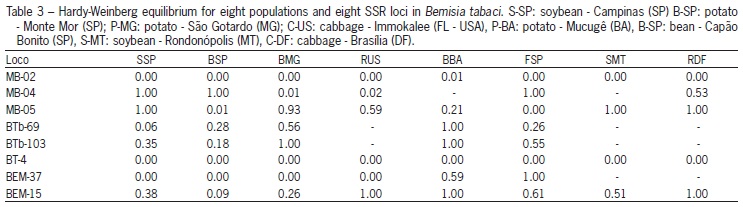
The populations are in Hardy-Weinberg equilibrium for most loci, except for MB02, BT-4 and BEM37 (p < 0.05) (Table 3). By the calculation of F statistics, the FIT value of 0.34 was significant, revealing that there is a factor acting on the populations in order to structure them. The estimate of FIS, which measures the deficiency of heterozygotes whithin populations did not differ from zero (p < 0.05), therefore not showing great influence on the distribution of variability of populations (Table 4).
The FST, which evaluates the degree of genetic structure among populations, had a moderate and significant value of 0.13 after sequential Bonferroni correction, showing that these populations are differentiating. The value of pairwise FST obtained between the populations (Table 5) indicated C-DF as the most divergent population, followed by B-SP and S-MT, with average B-SP values of 0.2, 0.17 and 0.16, respectively. In contrast, S-SP and P-SP had the lowest FST values (average of 0.11 and 0.12, respectively), indicating they are the less divergent than the others.
In the three populations of the insect from the State of São Paulo ocurring in common bean in Capão Bonito (B-SP, 24°00'21" S; 48°20'56" O), potatoes in Monte Mor (P-SP, 22°56'49" S; 47°18'57" O) and soybean in Campinas (S-SP), high FST values were obtained between the populations of Capão Bonito and Monte Mor, and Capão Bonito and Campinas, 0.13 and 0.12, respectively. On the other hand, the population of Campinas and Monte Mor had a reduction in the allele fixation index (0.05), suggesting that these two populations are not isolated from each other. The narrowing between these two populations is facilitated by geographical location, considering that Monte Mor and Campinas cities are geographically more close (30 km), than Capão Bonito, with is located approximately 200 km away from the two. But when geographic distance is regarded as a significant causative factor for population divergence, it does not always explain the structure presented. The whitefly population coming from São Gotardo (State of Minas Gerais, 19°18'39" S; 46°02'56" O) is 600 km away from the population of Monte Mor (SP) and the FST index between them is only 0.07. Another plausible explanation for the discrepancy observed between the whiteflies from Capão Bonito compared to those materials from Monte Mor and Campinas, is large-scale use of insecticides in bean plants in Capão Bonito, where selection pressure could be causing a local adaptive effect, hence changing gene frequencies of the population, as suggested by De Barro (2005). It should also be taken into account the presence of host-plant factors which, by exerting a selection pressure on the colonizing population, causes allele frequencies to modify according to the need for adaptation in the culture. In fact, field observations show that the bean plant is not considered a good host for B. tabaci when compared with other plants such as soybean, cabbage, tobacco and eggplant for their potential to rear whitefly.
The whitefly populations in potato crops had lower FST values when compared to other populations in other cultures (0.12, for P-SP and 0.13 for P-BA and P-MG). In general, even though the populations in potato have revealed low genetic structure, the FIS values that measure inbreeding whithin subpopulations, were not the highest compared to all other populations (Table 4), with the values of 0.23, 0.21 and 0.16 for P-BA, P-SP and P-MG, respectively. This result was not expected because being a relatively recent host of the pest it was more appropriated to have a high level of homozygotes in this populations due to inbreeding, if were assumed that the appearance of this pest in potato started from few individuals.
The population of whiteflies captured in cabbage crops in the U.S. (C-US), which should be very different from the Brazilian populations due to greater geographical distance, presented mostly intermediate FST values [average value of 0.13, minimum of 0.08 (BSP) and a maximum of 0.20 (RDF)], corroborating with the idea that the B biotype could probably be introduced from the U.S. into the region of Campinas (Lourenção and Nagai, 1994).
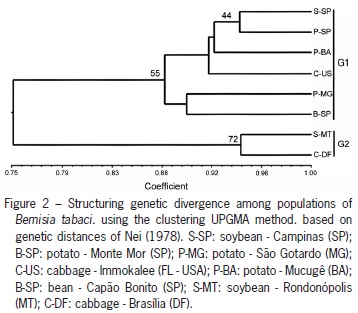
Genetic distances obtained among populations, which ranged from 0.06 (S-SP X P-SP) to 0.43 (B-SP X C-DF) were employed to generate the dendrogram (UPGMA) (Figure 2). Two main groups were identified, and populations in the midwest of the country (G2) presented themselves as genetically distinct from the populations of other regions (G1). Genetic distances between populations (excluding the U.S. population) were not correlated with their geographical distances by the Mantel test. Although not significant (r = 0.36 and p = 0.13), we determined that the greatest genetic distances were obtained between geographically distant populations, i.e., P-SP / S-SP and S-MT / C-DF.
Using the Bayesian approach performed by the program STRUCTURE, we tested values from 2 to 10 for K populations, according to Evanno et al. (2005), and the optimal value of K was 2, with almost all individuals presenting Q > 0.8 (Figure 3). The genetic structure revealed by the Structure Program is related to the result obtained using the distance of Nei (1978). For the Bayesian approach, the two populations in the midwest (red) remained grouped with S-SP and were separated from B-SP, P-MG and C-US (green). Populations P-SP and P-BA share individuals in both groups, again showing the diversity found in insects colonizing potato fields. About not congruent grouping of individuals belonging to populations P-SP and P-BA in the analysis obtained from Structure and UPGMA, the answer is probably that the genetic structure between these two groups is not strong enough to keep them together in both analysis, which take into account different mathematical calculations.
The PCA plans represented by the main coordinates were obtained using the distance of Nei (1978). The genetic structure obtained in individuals of all studied populations according to components 1 and 2 produced cumulative variance of 53.8 % from the total (Figure 4). There appears to be a wide genetic divergence between whitefly individuals on all the eight SSR loci analyzed, where the FIT value found was 0.34, and significant. The most divergent populations whose FST values were higher (C-DF, B-SP and S-MT) were the most geographically isolated, which might explain the observed genetic structure (Figure 4).
The structure of genetic diversity of whitefly populations in this study is often interpreted based on the introduction of the B biotype insect in Brazil. The first report of B. tabaci was in 1923 (Bondar, 1928), in the State of Bahia, and this endemic population was related to biotype A. Only at the end of the 1960's, outbreaks occurred in populations in southern Paraná and São Paulo States (Costa et al., 1973) and the introduction of the B biotype in the country was evidenced by substantial increases in insect populations in the early 1990's, reported in the State of São Paulo (Lourenção and Nagai, 1994).
The previously existing populations were out-competed by the B biotype that, with a higher fertility rate than its predecessor, came to displace the first one. Nowadays, biotype A is rarely found in Brazilian areas of intense agriculture. Fontes et al. (2010) could not detect its presence in the analysis using RAPD markers in cultures in Mato Grosso, Distrito Federal and São Paulo. Rocha et al. (2011) also found only the B biotype in the State of São Paulo evaluating 17 populations of whitefly in relation to the mtCOI sequence in crops like pepper, tomato, eggplant and Cucurbitaceae. However, it is possible that in some locations, such as in the northeast and midwest, the arrival of B biotype has occurred latter and in smaller populations and that, once in sympatry, enabled the occurrence of introgression between biotypes A and B, which would explain greater genetic similarity found between C-DF and S-MT and the distinction of these two populations in relation to others, for presenting allele frequencies more similar or greater number of alleles in common.
Fontes et al. (2010) used RAPD markers for these same populations in the midwest, also noted a differential clustering of midwest populations compared to others in the State of São Paulo, reinforcing that there is a high genetic structure. However, the hypothesis discussed above has several serious flaws since studies show that the different genetic groups (called biotypes) of the B. tabaci complex do not cross (Perring et al., 1993; Bedford et al., 1994), but display courtship behaviour (Perring and Symmes, 2006) and the rare observations of mating between biotypes (e.g. the B and AN) resulted in fewer offspring or infertile hybrids (DeBarro and Hart, 2000). Therefore, if the possibility of interbreeding between biotypes is disregarded, one could speculate that genetic differentiation between populations found in central Brazil (Distrito Federal and Mato Grosso) and the others is the result of the introduction of the B biotype from other places not analyzed in this study, or also due to the bottleneck effect, with anthropogenic dispersal from the insect's original center of introduction in Brazil in the central region of the country (Fontes et al., 2010).
The molecular markers employed have an important role due to their ability to quantify the genetic diversity and to differentiate groups of insects which are apparently morphologically identical but may greatly differ in their ability to vector viruses and cause crop damage. In this context, the results of this study have identified two major genetically distinct groups, represented by the populations of the central region of the country, which are genetically different to populations in other regions. Other differences may be associated with this genetic structure, such as preferential host, degree of dispersion, prolificacy, resistance to certain groups of insecticides, and ability to transmit viruses. These differences should be better explored in order to increase the efficiency of chemical or biological control of insect pests which vector viruses.
The great similarity between the US and mainly the SP populations support the hypothesis that B biotype was introduced into Brazil at some time probably by the trade of plant material from the US to the State of São Paulo. Moreover, insects coming from bean crops in Capão Bonito presented themselves genetically differentiated from the others, even of those from the same state. These results may be further evidence for the need to develop alternative, integrated management practices for insect control, considering that bean crops in this location are characterized by the overuse of insecticides, which can promote the development of increasingly resistant individuals, which is a characteristic of the B biotype (De Barro, 1995).
The magnitude of the genetic diversity observed among individuals in the southeast is greater than the diversity found in other regions, suggesting that this region was the most likely to the B biotype in Brazil and, probably, due to its intensive and very differentiated agriculture, remains the largest center of genetic diversity of this pest in the country, as shown by Lourenção and Nagai (1994).
Acknowledgements
To Dra. Geni Litvin Villas Boas and Dr. Philip Stansly for providing the whiteflies from Distrito Federal and from Florida (USA), respectively. To the Foundation of Research Support in São Paulo (FAPESP) for the scholarship granted to the first author and to the National Council of Scientific and Technological Development (CNPq) for productive research scholarship granted to the third author. To INCT interactions Plant-Pest.
Edited by: Richard V. Glatz
Received November 25, 2010 Accepted September 06, 2011
- Aljanabi, S.M.; Loiácono, M.S.; Lourenço, R.T.; Borges, M.; Tigano, M.S. 1998. RAPD analisys revealing polymorphism in egg parasitoids of soybean stink bugs (Hemiptera: Pentatomidae). Anais da Sociedade Entomológica do Brasil 27: 345-352.
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. 1994. Geminivirus transmission and biological characterization of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Annals of Applied Biology 125: 311-325.
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. 1996-2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Available at: http://www.genetix.univ-montp2.fr/genetix/genetix.htm [Accessed Dec. 10, 2009] (in French).
- Bondar, G. 1928. Aleyrodideos of Brazil. Boletim do Laboratório de Pathologia Vegetal do Estado da Bahia 5: 1-37 (in Portuguese).
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. 1995. The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annual Review of Entomology 40: 511-534.
- Byrne, D.N.; Bellows Jr, T.S. 1991. Whitefly biology. Annual Review of Entomology 36: 431-457.
- Costa, A.S.; Costa, C.L.; Sauer, H.F.G. 1973. Outbreak of whitefly in crops of Parana and Sao Paulo. Anais da Sociedade Entomológica do Brasil 2: 20-30 (in Portuguese, with abstract in English).
- De Barro, P.J. 1995. Bemisia tabaci biotype B, a review of its biology, distribution and control. CSIRO Division Entomology Technical Paper 36: 1-58.
- De Barro, P.J.; Hart, P.J. 2000. Mating interactions between two biotypes of the whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) in Australia. Bulletin of Entomological Research 90: 103-112.
- De Barro, P.J.; Scott, K.D.; Graham, G.C.; Lange, C.L.; Schutze, M.K. 2003. Isolation and characterization of microsatellite loci in Bemisia tabaci Molecular Ecology Notes 3: 40-43.
- De Barro, P.J. 2005. Genetic structure of the whitefly Bemisia tabaci in the Ásia-Pacific region revealed using microsatellite markers. Molecular Ecology 14: 3695-3718.
- De Barro, P.J.; Trueman, J.W.H.; Frohlich, D.R. 2005. Bemisia argentifolii (Hemiptera: Aleyrodidae) is a race of B. tabaci: the molecular genetic differentiation of B. tabaci populations around the world. Bulletin of Entomological Research 95: 193-203.
- De Barro, P.J.; Liu, S.; Boykin, L.M.; Dinsdale A.B. 2011. Bemisia tabaci: A Statement of Species Status. Annual Review of Entomology 56: 1-19.
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. 2010. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidae: Aleyrodidae) mitochondrial cytochrome oxidase I to identify level genetic boundaries. Annals of the Entomological Society of America 103: 196-208.
- Evanno, G.; Regnaut, S.; Goudet, J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611-2620.
- Fontes, F.V.H.M.; Colombo, C.A.; Lourenção, A.L. 2010. Molecular characterization and genetic divergence of Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae) on different crops and growing areas. Neotropical Entomology 39: 221-226 (in Portuguese, with abstract in English).
- Jones, D.R. 2003. Plant viruses transmitted by whiteflies. European Journal of Plant Pathology 109: 195-219.
- Kwok, S.; Kellog, D.E.; McKinney, N.; Spasic, D.; Goda, L.; Levenson, C.; Sninsky J.J. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus 1 model studies. Nucleic Acids Research 18: 999-1005.
- Lima, L.H.C.; Návia, D.; Inglis, P.W.; Oliveira, M.R.V. 2000. Survey of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidade) biotypes in Brazil using RAPD markers. Genetics and Molecular Biology 23: 1-5.
- Lima, L.H.C.; Campos, L.; Moretzsohn, M.C.; Návia, D.; Oliveira, M.R.V. 2002. Genetic diversity of Bemisia tabaci (Genn.) populations in Brazil revealed by RAPD markers. Genetic and Molecular Biology 25: 217-223.
- Lourenção, A.L.; Nagai, H. 1994. Outbreaks of Bemisia tabaci in the São Paulo State, Brazil. Bragantia 53: 53-59 (in Portuguese, with abstract in English).
- Mantel, N.A. 1967. The detection of disease clustering and a generalised regression approach. Cancer Research 27: 209-220.
- Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583-590.
- Oliveira, M.R.V.; Henneberry, T.J.; Andreson, P. 2001. History, current status and collaborative research projects for Bemisia tabaci Crop Protection, 20: 709-723.
- Perring, T.M.; Cooper, A.D.; Rodriguez, R.J.; Farrar, C.A.; Bellows, T.S.J. 1993. Identification of a whitefly species by genomic and behavioral studies. Science 259: 74-77.
- Perring, T.M.; Symmes, E.J. 2006. Courtship behavior of Bemisia argentifolii (Hemiptera: Aleyrodidae) and whitefly mate recognition. Annals of the Entomological Society of America 99: 598-606.
- Pritchard, J.K.; Stephens, M.; Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
- Pritchard, J.K.; Wen, W. 2004. Documentation for STRUCTURE software. The University of Chicago Press, Chicago.
- Raymond, M.; Rousset, F. 1995. Genepop version 2.0: population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248-249.
- Rice, W.R. 1989. Analysing tables of statistical tests. Evolution 43: 223-225.
- Rocha, K.C.G.; Marubayashi, J.M.; Navas-castillo, J.; Yuki, V.A.; Wilcken, C.F.; Pavan, M.A.; Krause-Sakate, R. 2011. Only the B biotype of Bemisia tabaci is present on vegetables in São Paulo State, Brazil. Scientia Agricola 68: 120-123.
- Rohlf, F.J. 1989. NTSYS-Pc: Numerical Taxonomy and Multivariate Analysis System. Exeter Publisher, New York, NY, USA.
- Simón, B.; Cenis, J.L.; De La Rúa, P. 2007. Distribution patterns of the Q and B biotypes of Bemisia tabaci in the Mediterranean Basin based on microsatellite variation. Entomologia Experimentalis et Applicata 124: 327-336.
- STATSOFT. Statistics for Windows.1999: Computer Program Manual]. StatSoft, Tulsa. OK, USA.
- Tsagkarakou, A.; Roditakis, N. 2003. Isolation and characterization of microsatellite loci in Bemisia tabaci (Hemiptera: Aleyrodidae). Molecular Ecology Notes 3: 196-198.
- Tsagkarakou, A.; Tsigenopoulos, C.S.; Gorman, K.; Lagnel, J.; Bedford, I.D. 2007. Biotype status and genetic polymorphism of the withefly Bemisia tabaci (Hemiptera: Aleyrodidae) in Greece: mitochondrial DNA and microsatellites. Bulletin of Entomological Research 97: 29-40.
- Weir, B.S. 1996. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer, Sunderland, UK.
- Weir, B.S.; Cockerham, C.C. 1984. Estimating F-Statistics for the analysis of population structure. Evolution 38: 1358-1370.
Publication Dates
-
Publication in this collection
16 Feb 2012 -
Date of issue
Feb 2012
History
-
Received
25 Nov 2010 -
Accepted
06 Sept 2011