Abstracts
Non-coding RNAs (ncRNAs) were recently given much higher attention due to technical advances in sequencing which expanded the characterization of transcriptomes in different organisms. ncRNAs have different lengths (22 nt to >1, 000 nt) and mechanisms of action that essentially comprise a sophisticated gene expression regulation network. Recent publication of schistosome genomes and transcriptomes has increased the description and characterization of a large number of parasite genes. Here we review the number of predicted genes and the coverage of genomic bases in face of the public ESTs dataset available, including a critical appraisal of the evidence and characterization of ncRNAs in schistosomes. We show expression data for ncRNAs in Schistosoma mansoni. We analyze three different microarray experiment datasets: (1) adult worms' large-scale expression measurements; (2) differentially expressed S. mansoni genes regulated by a human cytokine (TNF-α) in a parasite culture; and (3) a stage-specific expression of ncRNAs. All these data point to ncRNAs involved in different biological processes and physiological responses that suggest functionality of these new players in the parasite's biology. Exploring this world is a challenge for the scientists under a new molecular perspective of host-parasite interactions and parasite development.
Schistosoma mansoni; non-coding RNAs; gene expression profile; genome; transcriptome
RNAs não codificadores (ncRNAs) têm sido recentemente objeto de atenção muito maior devido aos avanços técnicos no sequenciamento que expandiram a caracterização dos transcritomas em diferentes organismos. ncRNAs possuem diferentes comprimentos (22 nt a >1.000 nt) e mecanismos de ação que essencialmente compreendem uma sofisticada rede de regulação de expressão gênica. A publicação recente dos genomas e transcritomas dos esquistossomos aumentou a descrição e caracterização de um grande número de genes do parasita. Aqui nós revisamos o número de genes preditos e a cobertura das bases do genoma em face dos ESTs públicos disponíveis, incluindo uma avaliação crítica da evidência e caracterização de ncRNAs em esquistossomos. Nós mostramos dados de expressão de ncRNAs em Schistosoma mansoni. Nós analisamos três conjuntos diferentes de dados de experimentos com microarranjos: (1) medidas de expressão em larga escala de vermes adultos; (2) genes diferencialmente expressos de S. mansoni regulados por uma citocina humana (TNF-α) no parasita em cultura; e (3) expressão estágio-especifica de ncRNAs. Todos estes dados apontam para ncRNAs envolvidos em diferentes processos biológicos e respostas fisiológicas que sugerem funcionalidade destes novos personagens na biologia do parasita. Explorar este mundo é um desafio para os cientistas sob uma nova perspectiva molecular da interação parasita-hospedeiro e do desenvolvimento do parasita.
Schistosoma mansoni; RNAs não-codificadores; perfil de expressão gênica; genoma; transcritoma
BIOMEDICAL SCIENCES
Non-coding RNAs in schistosomes: an unexplored world
Katia C. OliveiraI; Mariana L. P. CarvalhoI; Vinicius Maracaja-CoutinhoI; João P. KitajimaII; Sergio Verjovski-AlmeidaI
IDepartamento de Bioquímica, Instituto de Química, Universidade de São Paulo, Av. Prof. Lineu Prestes, 748, 05508-000 São Paulo, SP, Brasil
IIHospital Israelita Albert Einstein, Av. Albert Einstein, 627, 05652-000 São Paulo, SP, Brasil
Correspondence to Correspondence to: Sergio Verjovski-Almeida E-mail: verjo@iq. usp. br
ABSTRACT
Non-coding RNAs (ncRNAs) were recently given much higher attention due to technical advances in sequencing which expanded the characterization of transcriptomes in different organisms. ncRNAs have different lengths (22 nt to >1, 000 nt) and mechanisms of action that essentially comprise a sophisticated gene expression regulation network. Recent publication of schistosome genomes and transcriptomes has increased the description and characterization of a large number of parasite genes. Here we review the number of predicted genes and the coverage of genomic bases in face of the public ESTs dataset available, including a critical appraisal of the evidence and characterization of ncRNAs in schistosomes. We show expression data for ncRNAs in Schistosoma mansoni. We analyze three different microarray experiment datasets: (1) adult worms' large-scale expression measurements; (2) differentially expressed S. mansoni genes regulated by a human cytokine (TNF-α) in a parasite culture; and (3) a stage-specific expression of ncRNAs. All these data point to ncRNAs involved in different biological processes and physiological responses that suggest functionality of these new players in the parasite's biology. Exploring this world is a challenge for the scientists under a new molecular perspective of host-parasite interactions and parasite development.
Key words:Schistosoma mansoni, non-coding RNAs, gene expression profile, genome, transcriptome.
RESUMO
RNAs não codificadores (ncRNAs) têm sido recentemente objeto de atenção muito maior devido aos avanços técnicos no sequenciamento que expandiram a caracterização dos transcritomas em diferentes organismos. ncRNAs possuem diferentes comprimentos (22 nt a >1.000 nt) e mecanismos de ação que essencialmente compreendem uma sofisticada rede de regulação de expressão gênica. A publicação recente dos genomas e transcritomas dos esquistossomos aumentou a descrição e caracterização de um grande número de genes do parasita. Aqui nós revisamos o número de genes preditos e a cobertura das bases do genoma em face dos ESTs públicos disponíveis, incluindo uma avaliação crítica da evidência e caracterização de ncRNAs em esquistossomos. Nós mostramos dados de expressão de ncRNAs em Schistosoma mansoni. Nós analisamos três conjuntos diferentes de dados de experimentos com microarranjos: (1) medidas de expressão em larga escala de vermes adultos; (2) genes diferencialmente expressos de S. mansoni regulados por uma citocina humana (TNF-α) no parasita em cultura; e (3) expressão estágio-especifica de ncRNAs. Todos estes dados apontam para ncRNAs envolvidos em diferentes processos biológicos e respostas fisiológicas que sugerem funcionalidade destes novos personagens na biologia do parasita. Explorar este mundo é um desafio para os cientistas sob uma nova perspectiva molecular da interação parasita-hospedeiro e do desenvolvimento do parasita.
Palavras-chave:Schistosoma mansoni, RNAs não-codificadores, perfil de expressão gênica, genoma, transcritoma.
BACKGROUND
Schistosomiasis is a parasitic debilitating disease widespread in the world. The disease occurs in 76 countries especially in America, Africa and Asia. It is estimated that 779 million people in these countries are at risk of this infection and around 207 million people are infected. Three major species of Schistosoma are responsible for the disease: S. mansoni (that occurs in America and Africa), S. japonicum (Asia) and S. haematobium (Africa and eastern Mediterranean) (Steinmann et al. 2006).
S. mansoni has a complex life cycle with six distinct developmental stages in two hosts: eggs, miracidia (first larval stage of free living), primary and secondary sporocists (inside the intermediate host - a snail), cercariae (second larval stage of free living, that infects the definitive host), schistosomula (inside the definitive host - a mammalian) and adult worms (after 42 days of infection) (Gryseels et al. 2006). The adult couple starts the oviposition process and migrates to the mesenteric veins. Eggs cross the epithelial layer of veins and intestinal wall and are eliminated in the feces, restarting the biological cycle; many eggs go through blood circulation and cause inflammatory processes especially in the liver and this is the main cause of the pathological process (Gryseels et al. 2006).
Schistosome couples can live for years inside the host, suggesting that they are completely adapted to the host environment. It is already known that schistosomes take advantage of host signals from endocrine and immune system, uptake nutrients for their development and differentiation (Amiri et al. 1992, De Mendonca et al. 2000, Davies et al. 2001, Escobedo et al. 2005, Han et al. 2009), and in addition the parasite has many ortholog genes to human receptors (Agboh et al. 2004, Osman et al. 2006, Khayath et al. 2007, Wu et al. 2007, Oliveira et al. 2009). It is also known that the parasite has a sophisticated alternative splicing mechanism for genes encoding secreted proteins such as micro-exon genes (MEGs) (Demarco et al. 2010), polymorphic mucins genes (SmPoMucs) (Roger et al. 2008) and venom allergen-like (SmVALs) genes (Chalmers et al. 2008). These mechanisms are supposed to increase the repertoire of parasite proteins (Verjovski-Almeida and Demarco 2011), possibly helping to evade the immune response. Understanding the molecular mechanisms that are responsible for such a diverse life cycle and that promote the sophisticated parasite's adaptation is a challenge to the research community.
OVERVIEW ABOUT THE CURRENT KNOWLEDGE ON NON-CODING RNA
In the last years with the advance of sequencing technologies, many genomes of model organisms as well as their transcriptomes have been sequenced and a huge amount of sequence information has become available to the scientific community (The_C.Elegans_Sequencing_Consortium 1998, Kaul et al. 2000, Carninci et al. 2003, Begun et al. 2007, Birney et al. 2007, Church et al. 2009). As a consequence of the analysis of these genomes, the central dogma of molecular biology, namely that genetic information flows from DNA to RNA to protein, the final effector in the cell, has been challenged (Mattick 2003). It has been observed that the majority of the mammalian genome is pervasively transcribed and different types of RNAs without protein-coding potential have been identified (Birney et al. 2007, Nakaya et al. 2007). Because of this new finding, nowadays it is much more complicated to define a gene. A current definition of gene is: "The gene is a union of genomic sequences encoding a coherent set of potentially overlapping functional products" (Gerstein et al. 2007).
In general, one protein-coding gene is defined by the presence of an ORF longer than 100 amino acids. It is estimated that up to 90% of the human genome is transcribed, however only 2% of these transcripts are protein-coding genes (Claverie 2005, Johnson et al. 2005, Birney et al. 2007). These data revealed that most of the transcripts are non-coding RNAs. In face of this scenario it has been argued that this may reflect transcription noise in the cell with no biological relevance (Huttenhofer et al. 2005, Werner and Berdal 2005). In the opposite scenario, important biological functions including regulatory networks that directly involve ncRNA molecules have been described in the last years (Mattick 2004, Reis et al. 2005, Dinger et al. 2009, Louro et al. 2009, St. Laurent et al. 2009, Chen et al. 2010a, De Lucia and Dean 2010, Nag and Jack 2010).
Distinct characteristics of ncRNAs lead to the proposal that they have important regulatory functions: (i) conservation of their promoters, splice junctions, exons, predicted structures, genomic positions and expression (less than in protein-coding genes, nevertheless conserved) (Trinklein et al. 2004, Washietl et al. 2005, Louro et al. 2009, Mercer et al. 2009), (ii) their dynamic expression and alternative splicing during differentiation (Rinn et al. 2007, Guttman et al. 2009), (iii) their altered expression in cancer and other diseases (Reis et al. 2004, Brito et al. 2008, Nana-Sinkam et al. 2009, Kumar et al. 2010), (iv) their association with a particular chromatin signature that is indicative of actively transcribed genes (Dindot et al. 2009, Guttman et al. 2009), (v) their regulation by key morphogens and transcription factors (Cawley et al. 2004, Li et al. 2010); (vi) their tissue and cell-specific expression pattern and sub cellular localization (Louro et al. 2007, Nakaya et al. 2007, Birney et al. 2007, Mattick 2009).
In our perspective, one of the most important points of this discussion is the fact that the complexity of organisms along the evolution has been associatedmparison
with the expansion of genomic elements. Co between the increasing number of protein-coding genes and non-protein coding genes clearly reveals that the expansion of ncDNA (especially the intronic regions) is much higher than the expansion of protein coding genes (Mattick 2004). It raises the hypothesis that regulation of complex functions is mediated by sophisticated mechanisms involving ncRNAs. Probably this was not noticed before because of the protein-centered view of molecular biology. Accumulation of large amounts of sequencing data (Core et al. 2008, Mortazavi et al. 2008, Oliver et al. 2009) and of tiling array experiments (Johnson et al. 2005, Wilhelm et al. 2008) was necessary to show pervasive transcription especially in higher eukaryotes. In consequence of these observations the number of articles related with the description and study of ncRNAs has increased considerably in the last years (Mattick 2009).
Presently many types of ncRNAs have been characterized and described in the literature. Short ncRNAs are the most studied class of ncRNA in humans and model organims; they include microRNAs which are small RNAs (22 nt long) that regulate gene expression of hundreds to thousands genes by partial complementary base pairing to specific mRNAs (a post-transcription regulation). They direct degradation of target mRNAs through cleavage by Argonaute enzyme (present in RISC complex) (Bartel 2004); and siRNAs, which are endogenous small RNAs with 21 nt length produced by Dicer cleavage of perfectly complementary dsRNA duplexes. They form complexes with Argonaute proteins and are involved in gene regulation, transposon control and viral defense; these RNAs can act in cis or in trans (Brosnan and Voinnet 2009). Long ncRNAs (lncRNAs) are defined as RNAs of little protein-coding potential, with a length higher than 100-200 bp (arbitrary limit). They are the least understood transcriptional unit and comprises a heterogeneous group of transcripts (Costa 2010); Intronic long ncRNAs can be the product of a splice processing or originate from an independent transcription (Rearick et al. 2010); Large intergenic ncRNAs appear to be selected for conservation by evolution and are associated with epigenetic regulation (Guttman et al. 2009).
Another important concept in non-coding RNA is related toNATs: Natural AntisenseTranscripts. These are generally non-protein coding transcripts, but fully processed, mRNAs that are transcribed from the opposite strand of protein-coding sense transcript (Werner and Swan 2010). Studies reveal a conservation of these transcripts among human, mouse and fish (Dahary et al. 2005, Zhang et al. 2006). These transcripts can act as precursor of siRNA, miRNA, gene silencing, although the roles of NATs are not completely understood.
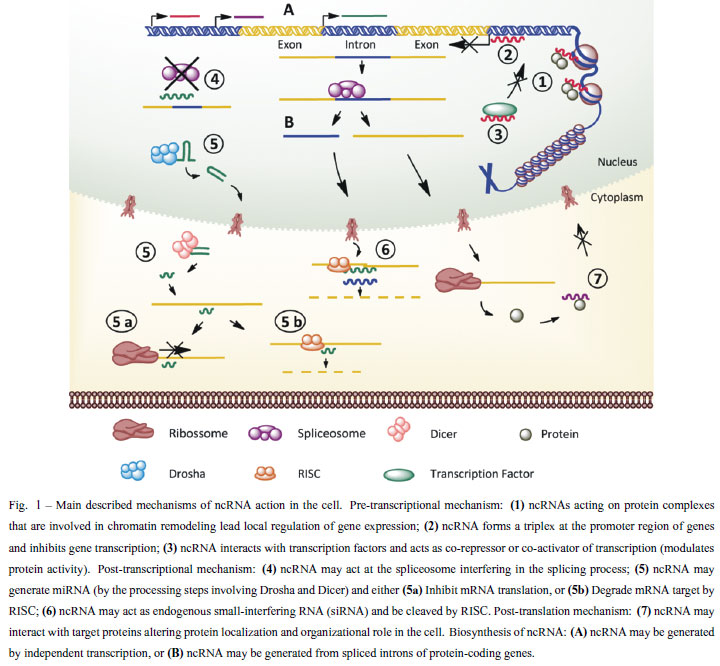
Different mechanisms are involved in the action of ncRNA; here we summarize in Figure 1 the described mechanisms in model organisms (Brosnan and Voinnet 2009, Mercer et al. 2009, Wilusz et al. 2009, Chen and Carmichael 2010).
THE GENOME AND TRANSCRIPTOME OF SCHISTOSOMES
The genome of schistosomes is organized in eight chromosomes, seven autosomal and one sexual. In 2003 the transcriptomes of S. mansoni (Verjovski-Almeida et al. 2003) and S. japonicum (Hu et al. 2003) were published in Nature Genetics, giving insights and perspectives for functional genomics (Verjovski-Almeida et al. 2004). Six years later, in 2009, the genome sequences from both parasites were published in Nature (Berriman et al. 2009, Zhou et al. 2009). Additionally, the genome sequencing project of a third Schistosoma species, S. haematobium, is on the way and will provide a new collection of sequences in a not too distant future (Webster
et al. 2010).
Table I summarizes the features of S. mansoni and S. japonicum published genomes (Berriman et al. 2009, Zhou et al. 2009) and all public EST transcripts. We can clearly see the similarities in the genome structure between these two schistosome species.
EST vs. GENE PREDICTIONS: HOW MANY S. mansoni GENES? HOW MANY POTENTIAL NON-CODING RNAS?
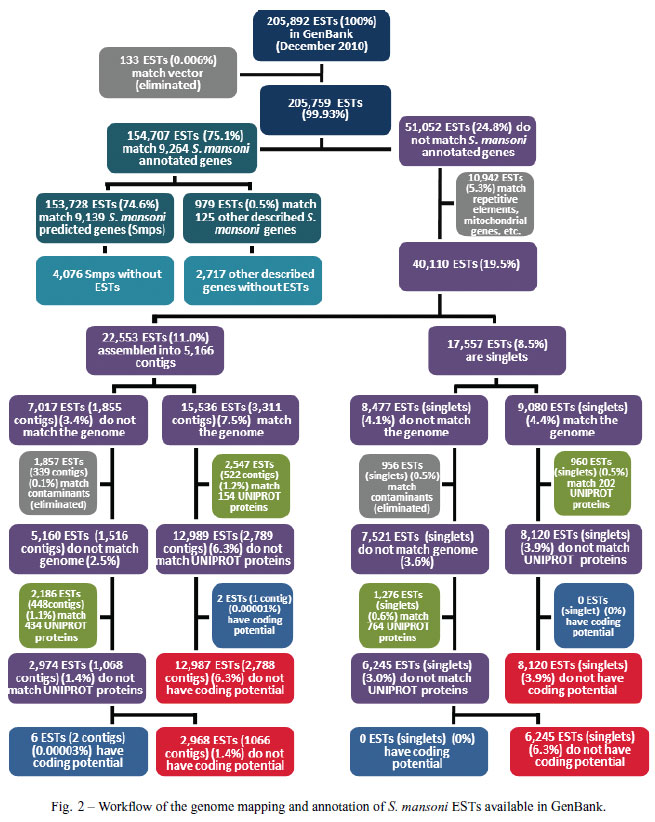
We performed a comparison between the transcriptome and genome of S. mansoni in order to determine the percentage of transcripts that may be related to potentially non-coding RNAs (Fig. 2). We used all 205,892 public S. mansoni ESTs and mRNAs available in Gen- Bank at the beginning of December 2010; in the first step of the analysis, we filtered out ESTs that match vectors (133 ESTs). Out of the remaining 205,759 ESTs we found that 154,707 (75.1%) (Fig. 2, upper part) could be mapped to S. mansoni annotated genes (i.e. 13,215 Smp protein-coding gene predictions (Berriman et al. 2009) plus 2,842 other described non-coding genes available at the Sanger Institute website (http://www.sanger.ac.uk/resources/downloads/helminths/schistosoma-mansoni.html), such as tRNAs, microRNAs, small nucleolar RNAs and ribosomal RNAs, which will
be discussed later in this review).
From the 51,052 ESTs that do not match Smps or other described genes, we filtered out 10,942 ESTs that match repetitive elements, mitochondrial genes, etc. The remaining 40,110 ESTs (19.5%) were assembled using CAP3 with default parameters, generating 5,166 contigs (comprised of 22,553 ESTs, 11.0%) and 17,557 EST singlets (8.5%) (Fig. 2).
The assembled ESTs described above were found to be divided into a major set that matches the genome outside of any predicted Smp gene (15,536 ESTs (7.5%) assembled into 3,311 contigs plus 9,080 singlets (4.4%); a total of 24,616 ESTs, 11.9%) and a smaller set that does not match the genome (7,017 ESTs assembled into 1,855 contigs plus 8,477 singlets; a total of 15,494 ESTs, 7.5%) (Fig. 2). Overall, our analysis shows that 87% of public S. mansoni ESTs match the genome and highlights the fact that a considerable fraction (11.9%) shows evidence of transcribed regions in the genome for which no Smp gene prediction was made (Berriman et al. 2009). Additionally, 4,076 Smps and 2,717 other genes were predicted in the genome without S. mansoni ESTs evidence.
We analyzed the protein coding potential among the 40,110 ESTs (19.5%) that do not match Smps. In a first step we looked for a match of the assembled ESTs to a curated protein dataset: UNIPROT (The_Uniprot_Consortium 2010) available at (http://www.uniprot.org/); in a second step, the assembled ESTs that did not match UNIPROT were analyzed for their protein-coding potential using Coding Potential Calculator (CPC) (Kong et al. 2007).
The ESTs that do not match Smps and match the genome (26,616 ESTs) were assembled into 3,311 contigs (15,536 ESTs, 7.5%) (Fig. 2) and we found that 522 of these contigs (2,547 ESTs, 1.2%) have match to 154 UNIPROT known proteins from other organisms and were not predicted in S. mansoni; one additional contig (composed of 2 ESTs, 0.00003%) that does not match UNIPROT was predicted by CPC to have a protein- coding potential (Fig. 2). The remaining 2,788 contigs (12,987 ESTs, 6.3%) are potential non-coding RNAs since they do not match UNIPROT proteins and were not predicted by CPC to have protein-coding potential. Out of the 9,080 EST singlets (4.4%) that match the genome outside of Smps, we found that 960 ESTs (0.5%) match 202 UNIPROT known proteins; the remaining 8,120 EST singlets (3.9%) are again potential non-coding RNAs since they do not match UNIPROT and were not predicted by CPC to have protein-coding potential.
Here we conclude that overall, 21,107 ESTs (10.3%) that match the genome have no protein-coding potential and are good candidates for S. mansoni noncoding RNAs; these ESTs point to 10,908 genomic regions (2,788 contigs + 8,120 EST singlets) with evidence of ncRNA transcription. These data also point to 356 known UNIPROT proteins (2,547 ESTs assembled into 522 contigs and 960 singlets that match genome) that were expressed in S. mansoni, map to the genome sequence and were not predicted by the genome project (see Supplementary Table I).
A total of 15,858 ESTs (7.7%) do not match the genome and are either transcribed from a non-sequenced part of the S. mansoni genome or represent contaminants in the transcriptome database, especially the EST singlets. Out of 1855 contigs (7,017 ESTs, 3.4%) we found that 339 contigs (1,857 ESTs, 0.1%) match contaminant sequences (such as M. musculus, bacteria, H. sapiens, B. glabata, B. taurus and R. norvergicus) (Fig. 2). Among the remaining 1,516 contigs (5,160 ESTs, 2.5%), 448 contigs (2,186 ESTs) match 434 UNIPROT proteins; 1,068 contigs (2,974 ESTs) do not match UNIPROT proteins. From this group, 2 contigs (6 ESTs) have coding potential and 1,066 contigs (2,968 ESTs, 1.4% of all transcripts) do not have protein-coding potential according CPC.
From the 8,477 EST singlets that do not match the genome we found that 956 have match to sequences of potential contaminants. From the remaining 7,521 singlets, 1,276 have match 764 UNIPROT proteins and 6,245 ESTs (3.0%) do not match UNIPROT proteins and do not have protein-coding potential (Fig. 2).
We could conclude that 9,213 ESTs (4.4%) between contigs and singlets that do not match the genome are potentially non-coding RNA. These data also point to 1,443 UNIPROT unique proteins that are conserved and are potentially expressed in S. mansoni but are not present in the genome (see Supplementary Table I).
In summary, among the 205,892 public S. mansoni ESTs a total of 30,320 ESTs (between contigs and singlets) (14.7%) do not have protein-coding potential; of these, 21,107 ESTs (10.2%) match the genome, while only 9,213 ESTs (4.5%) do not match the genome. The fraction of total S. mansoni transcription comprised by non-coding RNAs probably reveals a lower level of transcriptional activity of this class of RNAs, compared to the transcriptional activity of protein-coding genes. In fact, it has been reported in humans that the long non-coding RNAs are transcribed at a much lower rate than the protein-coding genes (Kapranov et al. 2007) and the non-coding RNAs are represented by between 10 and 20% of the human EST database collection (Nakaya et al. 2007).
COVERAGE OF ESTS ONTO THE GENOME AND GENE PREDICTIONS: A SURPRISING TRANSCRIPTION FROM THE INTRONS?
We calculated the percentage of bases in the genome that is comprised of gene predictions and the number of bases in the genome that are covered by NCBI public ESTs. The genome of S. mansoni has 362,876,148 bp distributed in 5,745 scaffolds >2 kbp (Berriman et al. 2009). From all these bases, 165,206,376 bp (45.5%) are loci of predicted genes. From these loci, 15,852,242 bp are exons of gene predictions (4.3% of total bases in the genome and 9.6% of gene prediction loci) and 149,354,134 bp (41% of total bases in the genome and
90.4% of gene prediction loci) are introns.
Based on the public S. mansoni ESTs that have so far been accumulated and that mapped to the sequenced part of the genome, we find that a total of 16,516,608 genomic bases were covered by at least one EST, which means that at least 4.6% of the S. mansoni genome is transcribed.
From the 16,516,608 transcribed base pairs, a total of 12,717,085 bp (77% of transcribed bases) is located in gene prediction loci (3.5% of genome bases). A total of 7.7% bases in gene prediction loci are covered; the loci include exons and introns of genes.
When looking at the exons in the genome (comprised of 15,852,242 bp), we found that 8,652,015 bp were covered by public ESTs (42% of transcribed bases) which represents coverage of 55% of exon bases of the predicted gene sequences (2.4% of genome bases); out of them 1,557,580 bases are in UTRs (Fig. 3). Looking at the predicted genomic introns (149,354,134 bp) we found that 4,065,070 bp were covered by public ESTs (34% of total transcribed bases) corresponding to coverage of only 2.7% of predicted intron bases (1.1% of genomic bases). We detected that 3,799,523 transcribed bases (1% of the total genomic bases and 23% of transcribed bases) are located in intergenic regions. Figure 3 summarizes these numbers.
Mattick (Mattick 2004) raised the hypothesis that the complexity of an organism probably is derived from the expansion of non-coding regions in the genome, especially because there is no considerable increase in the number of protein-coding genes along the evolution, whereas there is an important expansion of non-coding regions in the genomes of the more complex organisms. This expansion occurred especially in the intronic regions of protein-coding genes. Mattick calculated the ratio between ncDNA/total DNA for a large spectrum of organisms with diverse complexities, and found that complex organisms such as Mus musculus and Homo sapiens had a value higher than 0.9 (Mattick 2004). Nevertheless, the position of, for example, Anopheles gambiae, does not fully concord with the hypothesis. Moreover, within-clade variations can be considerable. For example in the ray-finned fishes, genome size (even allowing for polyploidy) can vary by a factor of 20-fold (Smith and Gregory 2009). Assuming that the number of coding genes is unlikely to vary much within this group, factors other than complexity may govern the amount of ncDNA in these genomes. In fact, factors such as metabolic rates, body size, effective population size are known to affect genome size (Keeling and Slamovits 2005).
Based on the genome and gene predictions (Berriman et al. 2009), here we calculated the ncDNA/Total DNA ratio in S. mansoni and found it to be 0.96, a quite high ratio considering the parasite complexity and comparing to the ratio in the human genome. In addition, S. mansoni has an unusual intron size distribution (Webster et al. 2010). It is interesting to note that another platyhelminth, the free-living Schmidtea mediterranea has a 480 Mb genome (http://genome.wustl.edu/genomes/view/schmidtea_mediterranea/), around 100 Mb bigger than S. mansoni and unlikely to have much more coding DNA, which would result in a ncDNA/Total DNA ratio even higher than S. mansoni; these observations suggest that the expansion of ncDNA may be one of the mechanisms used by evolution to achieve platyhelminth complexity and to interact with the environment. S. mansoni may have undergone genome reduction (Keeling and Slamovits 2005) in comparison to S. mediterranea due to parasitism of the former.
So far, there is limited evidence of transcriptional activity along the S. mansoni genome; just 4.6% of all genome bases are covered by EST data, as described above. The predicted intronic regions in S. mansoni are large; they comprise 41% of total genomic bases and 90% of genomic loci of gene predictions; in S. mansoni, transcription detected in intronic regions corresponds to only 1.1% of total genomic bases although it reveals that 34% of the transcribed bases are in introns. In humans 30% of the genome is comprised of introns and a pervasive transcription has been detected (Birney et al. 2007, Kapranov et al. 2007). In humans, an analysis of the 5.3 million public ESTs pointed to the presence of at least one EST mapping to the introns of 74% of all RefSeq human genes (Nakaya et al. 2007). In fact, we re-analyzed the genome mapping of 8 million public human ESTs, and we found approximately 70,000 unique intronic loci covering nearly 42 million genomic bases (1.7% of the human genome) with evidence of transcription.
An intensive effort has been placed in the study of intronic transcription of a segment corresponding to 1% of the human genome by a network of laboratories under the name of ENCODE Project, using different methods such as tiling-arrays, RNA-seq and paired-end sequencing; ENCODE found that 93% of the studied 1% segment was transcribed; both the intronic and intergenic regions showed evidence of transcription (Birney et al. 2007), and the suggestion is that this figure can be extrapolated to the entire human genome. In other higher eukaryotes such as C. elegans 70% of the genome is transcribed and an ENCODE analysis is being developed (Gerstein et al. 2010). In D. melanogaster 85% of the genome is transcribed (Graveley et al. 2010).
Given the above numbers, it is not surprising that that 34% of the transcribed bases in the public S. mansoni EST database are in introns. Nevertheless, the limited evidence of genomic coverage of intronic transcription in S. mansoni (only 1.1% of total genomic bases) can be explained by the fact that until now a limited number of sequencing projects were executed (Franco et al. 1995, 2000, Merrick et al. 2003,Verjovski-Almeida et al. 2003); in addition, the sequencing projects in S. mansoni were performed with poly-A transcripts, and it is known that many transcripts, especially non-coding RNAs are not poly-adenylated (Kiyosawa et al. 2005). It suggests that a big effort towards deep-sequence and tilling array approaches is warranted to obtain higher transcription coverage of the S. mansoni genome.
NON-CODING RNAS AND SCHISTOSOMES
SHORT NON-CODING RNAS
The S. mansoni transcriptome project (Verjovski-Almeida et al. 2003) provided data for the identification of several ESTs encoding proteins related to components of the microRNA processing machinery. Various papers have been published in recent years; most of these published works are related to the identification of the RNAi/miRNA pathway as well as to individual
miRNA identification, as detailed below.
The current model of RNAi processing involves two cleavage steps, each one centered on a ribonuclease enzyme. The precursor RNA (either a dsRNA or a miRNA primary transcript) is processed into a short inhibitory RNA (siRNA) by RNAse III enzymes called Drosha (in the nucleus) and Dicer (in the cytoplasm), with dsRNA binding domain (dsRBD) protein acting as a cofactor. In the second step, siRNA is loaded into the effector protein complex called RNA-induced silencing complex (RISC). This siRNA is opened in a strand-specific manner during RISC assembly and single-stranded siRNA locates its cognate mRNA target by base pairing. Gene silencing is the result of the nucleolytic degradation of the RNAse H enzyme Argonaute. If the siRNA/mRNA duplex contains mismatches at the scissile site, as is often the case of miRNAs, the mRNA target is not cleaved and gene silencing results from translational inhibition (Pratt and Macrae 2009).
The complete conserved machinery to process microRNA or siRNA has been identified in S. mansoni and S. japonicum. In 2008 Krautz-Peterson and Skelly (Krautz-Peterson and Skelly 2008a) described the Dicer gene in S. mansoni, and observed the highest expression levels in schistosomula (15 days-old) and eggs. Later, in 2009 Gomes et al. (Gomes et al. 2009) described Dicer, Drosha and four different Argonaute proteins in S. mansoni; they also observed the highest expression level in eggs, but not in schistosomula.
In S. japonicum, two articles were published in 2010; Chen et al. (Chen et al. 2010b) described three Argonaute proteins and observed the highest expression levels in eggs and miracidia; and Luo et al. (Luo et al. 2010) described Dicer and four Argonaute proteins. They also observed that the highest expression levels of Dicer and Argonaute 1 are in eggs and miracidia.
In general these authors are in agreement in their observations and this pattern of expression suggests that the miRNA regulatory pathway might take part in the transformation and development of schistosomes. Another hypothesis is that miRNA could be responsible for the repression of translation in eggs (Schier 2007).
The characterization of miRNA pathway genes points to the mechanism that causes RNA interference in schistosomes. The RNAi approach was already used in several areas of S. mansoni biology (Boyle et al. 2003, Skelly et al. 2003, Correnti et al. 2005, Delcroix et al. 2006, Dinguirard and Yoshino 2006, Freitas et al. 2007, Krautz-Peterson et al. 2007, 2010, Ndegwa et al. 2007, Krautz-Peterson and Skelly 2008b, Morales et al. 2008, Pereira et al. 2008, Faghiri and Skelly 2009, Rinaldi et al. 2009, Beckmann et al. 2010) as well as in S. japonicum (Cheng et al. 2005, Zhao et al. 2008, Kumagai et al. 2009, Zou et al. 2010). RNAi has proved itself as an important tool to elucidate gene function in schistosomes, in a similar way as in other organisms.
RNAi experiments have mostly used the electroporation method to deliver the dsRNA or RNAi into the parasite, with soaking or biobalistic delivery being also used. However, the mechanism of dsRNA uptake used by the worm remains unclear. Recently Krautz- Perterson (Krautz-Peterson et al. 2010) described a S. mansoni homolog protein to SID-1 (Systemic RNA Interference- Defective) of Caenorhabditis elegans, a multi-membrane spanning RNA importing protein, that contains 21 exons and potentially encodes a protein with 1018 amino acids. This SID-1 protein has been shown to be required for uptake of dsRNA in C. elegans (Winston et al. 2002); probably SmSID-1 may have the same function in S. mansoni. Localization of this transport protein on the parasite and its functional characterization are some interesting subjects that remain to be clarified.
The first miRNAs characterized in schistosomes were described in 2008 (Xue et al. 2008). In that work the authors described 227 cloned microRNAs in S. japonicum. Among the cloned miRNAs, five have high level of conservation with well characterized microRNAs in more complex organisms, such as human, mouse, C. elegans, Drosophila: let-7, miR-71, miR-new1, mir-125 and bantam.
Huang et al. (Huang et al. 2009) described 176 miRNAs in S. japonicum (including let-7, miR-71, bantam and miR-125), among them 172 novel miRNAs. All these new miRNAs were identified and mapped to the genome by the presence of an inferred RNA hairpin with pairing characteristics of known miRNA structure. The authors also analyzed the differential expression between mixed adult worms and hepatic schistosomula and observed that 35 out of 176 were expressed in adult worms, 60 in schistosomula and 81 in both stages.
In 2010 two more publications focused on the identification and characterization of short ncRNAs in S. japonicum using deep-sequencing approach (Hao et al. 2010, Wang et al. 2010); this strategy has proved itself to be a powerful technique to identify small ncRNAs.
Wang et al. (Wang et al. 2010) described 20 species- conserved miRNAs and 16 schistosome-specific miRNAs. These miRNAs were validated using northern blot or stem-loop qRT-PCR approaches. The paper also described the identification of 4,858 putative endogenous siRNAs, 40% of them related to retrotransposons (TE-derived) (Wang et al. 2010) as expected by comparison to reports from other species (Golden et al. 2008).
Hao et al. (Hao et al. 2010) sequenced 5.3 and 4.2 million reads from small RNAs from adult worms and schistosomula respectively; these sequences represent around 1.1 million unique clean sequences. In both stages, the majority of sequences are siRNAs, and they are Transposable-Elements-derived. The authors point to the identification of 38 unique S. japonicum transcripts and 16 miRNAs that belong to 13 miRNA families conserved in other metazoan organisms. The amount of siRNAs was at least 4.4 times larger in schistosomula and 1.6 times larger in adult worms than in other stages.
More recently, Simoes et al. (Simoes et al. 2011) performed a bioinformatics homology-based analysis and identified conserved miRNA in S. mansoni. The authors also identified 211 novel miRNA in S. mansoni by sequencing of small-RNA cDNA libraries from adult worms. Out of these 211 candidates, 11 miRNAs had their expression level validated by northen blot analysis; three out of these miRNAs were already described in S. japonicum.
OTHER NON-CODING RNAS
In 1998 Ferbeyre et al. (Ferbeyre et al. 1998) performed an in silico search for RNA structural motifs in sequence databases and have found a hammerhead ribozyme domain encoded in the satellite repetitive DNA of S. mansoni (Ferbeyre et al. 1998). Transcripts are expressed from these repeats as long multimeric precursor RNAs that cleave in vitro and in vivo into unitlength fragments. This RNA domain is able to engage in both cis and trans cleavage typical of the hammerhead
ribozyme (Ferbeyre et al. 1998).
Copeland et al. (Copeland et al. 2009) carried an extensive in silico search in the genome of S. mansoni and S. japonicum and performed a homology-based annotation of the "house-keeping" ncRNAs in schistosomes. The authors were able to identify 23 types of ncRNAs with conserved primary and secondary structure; among these we mention rRNA, snRNA, SLRNA, SRP, tRNA and RNase P, and possibly MRP and 7sK RNAs. The previously described hammerhead ribozyme RNA (Ferbeyre et al. 1998) is the most diverse because it originates from repetitive DNA; tRNAs were found to be the next most diverse ncRNAs encoded in the S. mansoni genome (tRNAscan-SE predicted a total of 713 tRNAs) (Copeland et al. 2009). The authors focused on the comparison between tRNA populations in other schistosomes and in a free-living platyhelminth organism (Schmidtea mediterranea); they also confirmed in S. mansoni the first miRNAs described in S. japonicum by Xue et al. (Xue et al. 2008).
Until now few articles studied the expression and characterization of ncRNAs in schistosomes, as reviewed above. Specifically, nothing can be found in the literature about long (>200 nt) ncRNAs.
DIFFERENTIAL EXPRESSION OF NON-CODING RNAS IN SCHISTOSOMES: OUR EXPERIENCE AND PERSPECTIVE
DETECTION OF TRANSCRIPTION FROM BOTH GENOMIC STRANDS IN THE SAME LOCUS IN S. mansoni ADULT WORMS
Our group in 2007 studied the transcriptome of S. mansoni adult worms using a microarray platform with 44 k oligonucleotide (60-mer) probes (Verjovski-Almeida et al. 2007); we detected 156 genome loci that were represented by probes on both genomic strands for which therewas evidence of expression from both probes. From these, 9 loci were selected for validation using strandspecific RT-qPCR and we validated 6 loci. These 156 loci may be sources of ancestral Natural Antisense Transcripts (NATs) (Werner and Swan 2010), that have not
been characterized so far.
The above paper (Verjovski-Almeida et al. 2007) was published before the availability of S. mansoni gene predictions to the scientific community at Schisto Gene- DB website (http://www.genedb.org/Homepage/Smansoni). Here we carefully performed a re-annotation of this array platform and mapped the oligonucleotide probes to S. mansoni gene predictions and proteins available in GenBank (nr). This re-annotation is available as Supplementary Table II. With the re-annotation we found that 108 out of 156 loci with expression from both genomic strands do map to gene predictions. From these 108 loci, 18 map on protein-coding exons and therefore only one strand in the pair is an ncRNA. Another 18 of them map to the UTR region and an additional 72 map to intronic regions (2 loci in the same gene prediction). All these 108 are candidates of NAT transcription (Werner and Swan 2010). Out of the remaining 48 loci that do not match S. mansoni gene predictions, 27 match conserved proteins in GenBank and again only one strand in the pair is an ncRNA. Finally, 21 still have no match proteins either in GenBank or gene predictions.
This finding is extremely interesting because here we point to the first potential NATs in schistosomes. The list with these 135 loci with evidence of NATs is available in Supplementary Table III. From the 6 loci with expression in both genomic strands, which were validated by strand-specific RT-qPCR (Verjovski-Almeida et al. 2007), we found that 4 are located in gene prediction loci (highlighted in Supplementary Table III). They are: Smp_174720, Smp_136110, Smp_096790 (all of them mapped to intronic regions), and Smp_194860 (mapped to an exonic region). Probes that have the same orientation of the coding message and map to intronic regions may be revealing novel non-predicted exons of that given protein-coding gene. An alternative explanation remains, in that these introns may be genomic loci of independent transcription or the intron spliced from the immature pre-mRNA may be processed as a precursor of ncRNA (i.e. miRNA). Evidence of transcription has been obtained from the opposite strand of proteincoding predicted genes, which certainly points to independent antisense transcriptional events. The future molecular characterization of these candidates should help in understanding the molecular mechanism of gene regulation in schistosomes.
NON-CODING RNAS WITH EXPRESSION CHANGES INDUCED BY HUMAN TNF-α
In December 2009 our laboratory published the ortholog gene of TNF-α receptor in S. mansoni and characterized the effect of human TNF-α on the parasite gene expression using a microarray platform of 44k oligonucleotide probes (Oliveira et al. 2009). Here, we re-annotated the 44k oligonucleotide array according to gene predictions that appear in the genome publication (Berriman et al. 2009) in order to highlight the differentially expressed probes that map to the opposite strand of known proteincoding
genes (potentially ncRNAs regulated by TNF-α).
Expression changes induced by treatment with human TNF-α had been detected in newly transformed 3 h-old schistosomula in culture (1 h treatment). A set of 755 probes had been identified with a statistically significant (q-value < 0.05) differential expression between TNF-α treated and control early schistosomula (Oliveira et al. 2009); with the re-annotation we conclude that 686 unique genes were affected.
Among these 686 genes, 564 match S. mansoni gene predictions, 32 match S. japonicum gene predictions, 69 have match to conserved proteins in GenBank, comprising a total of 667 known coding genes, and 21 have no match. Among these 667 genes, 65 have significant changes in the expression level of the antisense message of the respective loci and 6 loci have significant expression changes in both sense and antisense messages. From the 6 loci with changes in the expression level in both strands, 3 of them have a decreased expression of sense and anti-sense messages in response to human TNF-α while the other 3 have a discrepant expression pattern; 3 of these 6 loci have pairs of probes that map to intronic regions of predicted genes, 2 inUTRregions and one in a coding exon. All the 65 gene loci with detected expression in the anti-sense strand and the 6 loci with expression in both genomic strands are listed in Supplementary Table IV (Part A).
In adult worms treated during 1 h or 24 h with TNF-α we had identified (Oliveira et al. 2009) two distinct expression patterns in treated adult worms: genes with transient expression changes (up-regulated at 1 h treatment and down-regulated at 24 h treatment, or the opposite pattern) and genes with sustained changes (upregulated at 1 h and 24 h treatment, or down-regulated throughout).
A set of 1,594 probes revealed statistically significant (q-value < 0.05) transient changes in expression (Oliveira et al. 2009). With the microarray re-annotation we conclude that there are 1404 unique genes with transient changes induced by TNF-α; 1048 genes have match to S. mansoni gene predictions, 54 match S. japonicum gene predictions, 203 match conserved proteins (GenBank), comprising a total of 1305 known proteincoding genes, and 99 have no match. Among these 1305 differentially expressed known protein-coding genes we verified that 177 coding genes have significant changes in the expression level of the anti-sense message from the respective loci, and 45 loci have expression changes in both sense and anti-sense messages. In the group of genes with differential expression in both loci strands, 28 of them have the same pattern of expression change in the sense/anti-sense pair of probes, while 19 genes have an opposite pattern of expression between sense and anti-sense probes.
In consequence of the re-annotation we observed that out of the 45 gene loci that have significant changes in expression in both strands, 29 have pairs of probes that map to intronic regions of predicted genes and 3 pairs of probes map to UTRs. All 45 loci with expression in both genomic strands and 177 gene loci with antisense expression are listed in Supplementary Table IV (Part B).
A group of genes had been identified with sustained changes in expression at 1 and 24 h TNF-α treatment (Oliveira et al. 2009). A total of 626 probes had a sustained change in expression; with the present microarray re-annotation we conclude that there are 584 differentially expressed unique genes with sustained changes in the expression pattern induced by TNF-α. From these 584 genes, 471 have match to S. mansoni gene predictions, 25 match S. japonicum gene prediction, 58 genes have match to conserved proteins in Gen- Bank, comprising a total of 554 annotated genes, and 30 have no match to GenBank. Among the 554 known coding genes, 7 gene loci have evidence of transcription in both strands (4 with expression induced by TNF-α in the messages from both strands and 3 with opposite expression pattern in each strand). Interestingly, in these 3 loci with opposite expression pattern between sense and anti-sense messages, the pair of probes maps to intronic regions of the protein-coding genes. We also observed in the group of 584 known protein-coding genes with sustained changes in the expression level that 3 genes have significant changes in the expression profile just in the anti-sense message of the locus; all these 10 gene loci with expression in anti-sense and in both strands are available in Supplementary Table IV (Part C).
Overall, the above data reveal a potential new set of 303 long non-coding RNAs in schistosomes that are anti-sense messages to known protein-coding genes whose expression is regulated by human TNF-α. Expression changes modulated by an exogenous regulatory molecule from the host (TNF-α) suggests some functionality for these ncRNAs; these long ncRNAs may participate in a sophisticated network of gene regulation in consequence of TNF-α signaling that deserves further characterization.
NON-CODING EXPRESSION SIGNATURE AMONG LIFE CYCLE STAGES
A number of papers already describe differences in gene expression among the developmental stages of schistosomes (Dillon et al. 2006, Vermeire et al. 2006, Jolly et al. 2007, Fitzpatrick et al. 2009, Gobert et al. 2009). Here we performed a set of experiments with 5 developmental stages (eggs, miracidia, cercariae, 7-dayold schistosomula and adult worms) using a 4k-element cDNA microarray platform that was designed to have a considerable fraction of probes for non-protein-coding genes (1133 probes); a detailed description of this platform is deposited in GEO under accession number GPL3929 (Demarco et al. 2006). This is the first microarray analysis of gene expression profile among life cycle stages that focuses on non-protein-coding genes in S. mansoni.
We analyzed two biological replica samples of each developmental stage; 3 ug amplified RNA (Wang et al. 2000) was labeled with Cy3 or Cy5 and hybridize to the arrays essentially as previously described (Demarco et al. 2006). The combination of samples on an array was: eggs vs. miracidia; cercariae vs. 7-day-old schistosomula and 7-day-old schistosomula vs. adult worms. We used a dye-swap approach to correct for any bias caused by dye incorporation or by intrinsic differential fluorescence yield of the dyes (Demarco et al. 2006). Raw data of this experiment is deposited in GEO under accession number GSE27026.
We used two different analyses approaches. In the first approach we identified differentially expressed genes between two consecutive developmental stages (eggs vs. miracidia; cercariae vs. 7-day-old schistosomula and 7-day-old schistosomula vs. adult worms) using SAM (Significance Analysis of Microarray) software (Tusher et al. 2001). Overall, we were able to find 1,423 differentially expressed genes between two developmental stages among all previously indicated comparisons (Table II). A detailed description follows below.
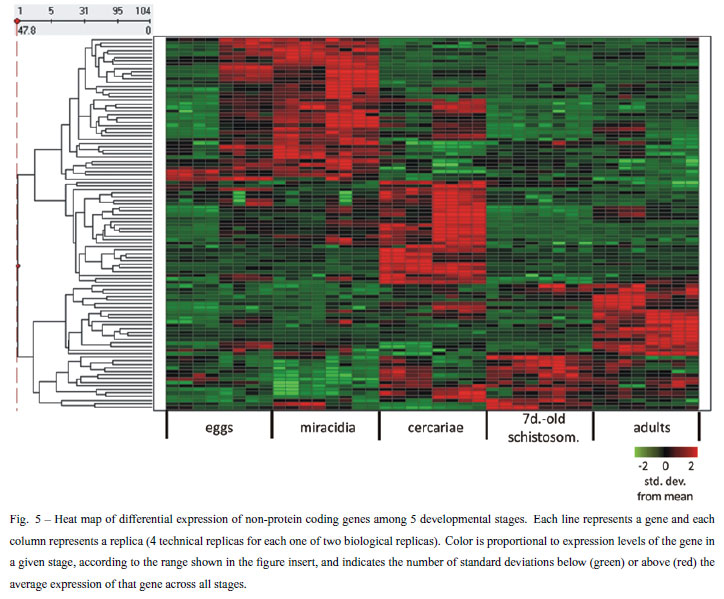
In the second approach we identified genes with increased expression levels in at least one developmental stage (for example more highly expressed in cercariae than in all other stages) using ANOVA statistical test (Churchill 2004) corrected for multiple sampling using Bonferroni correction (Shaffer 1995). Overall, with this approach we identified 577 differentially expressed genes with increased expression in at least one specific stage (Table III). A description of affected genes for each stage is given below.
In general, all observed patterns of expression of protein-coding genes among life cycle stages that will be described here, are not inconsistent with the previously published microarray results (Dillon et al. 2006, Jolly et al. 2007, Fitzpatrick et al. 2009, Gobert et al. 2009). Here we would like to especially highlight the non-coding genes that were never the focus of study before.
Differentially expressed non-coding genes identified here were mapped to the genome (using SchistoDB, available at http://schistodb.net/schistodb20/) and annotated as mapping to Intronic, Intronic/Exonic, Intergenic regions or as "Not mapped" (in case of multiple hits or no hit to the genome) (Tables II and III). These noncoding genes were re-confirmed as having no proteincoding potential by using the CPC tool (Kong et al. 2007). In addition, some previously annotated non-coding genes were now mapped to exons of predicted genes; they received an Smp re-annotation and were no longer counted as non-coding.
We are not able to assign a genomic strand for the observed expression of non-coding genes, since the probes on the 4k-microarray platform were generated by PCR amplification of selected double-stranded cDNA clones from the S. mansoni EST sequencing project (Verjovski-Almeida et al. 2003). These probes detect expression on either strand of a given locus.
Using the first analysis approach described above, we were able to find a set of 753 differentially expressed genes between eggs and miracidia; out of them, 117 genes with higher expression in egg and 636 genes with the opposite pattern. The complete list of differentially expressed genes between eggs and miracidia are available in Supplementary Table V (Part A). Among the 753 genes, there were 656 protein-coding genes (104 genes in eggs and 552 in miracidia); we highlight that there was significant enrichment (according to Gene Ontology analysis) of genes involved in amino acid and RNA metabolism in miracidia. The GO results are summarized in Supplementary Table V (Part B). We observed a set of 97 non-coding genes; 13 with higher expression in eggs and 84 with higher expression in miracidia (Table II). The non-coding expression profile is represented in Figure 4A. Description of genomic mapping coordinates is available in the supplementary material.
In the comparison between cercariae and 7-day-old schistosomula we found 401 differentially expressed genes; 271 in cercariae and 130 in schistosomula (Table II). From the 271 with higher expression in cercariae, 219 are protein-coding genes, and 52 are non-coding genes. Here, it is interesting to note the expression of a message that maps to the intronic region of the Smp_15-4340 gene, annotated as "nuclear factor Y transcription factor subunit B homolog, putative". This gene has three isoforms, and the intronic transcript could eventually act in cis modulating the splicing pattern of this transcript.
From the 130 genes with higher expression in 7-day-old schistosomula, 117 genes are protein-coding genes and 13 are non-protein coding (Table II). Figure 4B illustrates this profile. The list of all differentially expressed genes between cercariae and schistosomula is in Supplementary Table V (Part C). In this comparison we have found enriched GO categories both in cercariae and schistosomula; they are listed in Supplementary Table V (Part D and E), respectively.
In the third comparison, 7-day-old schistosomula vs. adults, we found 269 differentially expressed genes, 86 in schistosomula and 183 in adult worms. From the 86 genes with higher expression in schistosomula, 75 are protein-coding genes and 11 non-coding genes. In the opposite scenario, comprising the group of genes with higher expression in adult we have 155 proteincoding genes and 28 non-coding genes. This non-coding expression profile is represented in Figure 4C. No enriched GO categories were found among the proteincoding genes. The list of all differentially expressed genes is in Supplementary Table V (Part F).
Here we point to a set of non-coding genes that may be involved in molecular mechanisms of transformation that occur in each step of S. mansoni development.
In the second analysis approach we looked for differentially expressed genes with enriched expression in at least one stage, with the strategy explained before. We found 577 differentially expressed genes (Table III). Among these genes we found 473 protein-coding genes, and 104 non-protein coding genes. All 577 differentially expressed genes are listed in Supplementary Table V (Part G); the non-protein-coding genes signature is represented in Figure 5. In this profile of non-proteincoding genes we highlight one transcript that maps to an intron of Smp_014290; the Smp_014290 protein-coding gene has 3 alternatively spliced isoforms and it is possible that the non-coding RNA transcribed from its intron eventually acts in alternative splicing modulation; to confirm this hypothesis further studies are necessary.
The signature of stage-enriched expression comprises a new set of transcripts that should receive special attention in the future. This set reveals new molecular targets of specific mechanisms involved in the biology of each stage that should be explored to understand the parasite complexity.
FINAL CONSIDERATIONS / PERSPECTIVES
This is the first review on non-coding RNAs in schistosomes in the literature. Very limited knowledge about schistosomes non-coding RNAs is available. A large further effort using deep-sequencing and tilling array approaches is necessary to finish the genome, to increase the amount of transcriptome data and to identify new non-coding RNAs, especially in S. mansoni that until now has been less studied than S. japonicum with the non-coding perspective.
Extending genome and transcriptome deep sequencing to invertebrate species other than the C. elegans and D. melanogaster model organisms may help to identify the classes of ncRNAs in the ancient species. In addition, functional studies are necessary to clarify the mechanisms involved in regulation of these ncRNA and their role in protein-coding gene expression regulation. Because of the peculiarities of schistosomes and the current limited ability to obtain transgenic parasites, characterization of the molecular mechanisms of ncRNA function in schistosomes will be a big challenge.
Here we showed experimental evidence of regulation of non-coding RNAs expression in different biological situations: adult parasites, parasite response to TNF-α host molecule and parasite life cycle stages. These data collections are important evidence of functionality of these non-coding RNAs, and a detailed further characterization of the mechanisms of action is needed. Understanding of the non-coding genes as new players in the biology of schistosomes will shed light on the complexity of processes involved in host-parasite interaction and parasite development.
ACKNOWLEDGMENTS
Funded in part by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and from the SEtTReND grant agreement number 241865 of the FP-7 European Community. KCO and VMC received fellowships from FAPESP; MLPC received a fellowship from CAPES-Brasil, and SVA received an established investigator fellowship award from CNPq.
SUPPLEMENTARY MATERIALS
All supplementary materials are available for download from the following site: http://www2.iq.usp.br/docente/verjo/downloads/oliveira_kc/2011/
Manuscript received on February 5, 2011; accepted for publication on April 28, 2011
- AGBOH KC, WEBB TE, EVANS RJ AND ENNION SJ. 2004. Functional characterization of a P2X receptor from Schistosoma mansoni J Biol Chem 279: 4165041657.
- AMIRI P, LOCKSLEY RM, PARSLOW TG, SADICK M, RECTOR E, RITTER D AND MCKERROW JH. 1992. Tumour necrosis factor alpha restores granulomas and induces parasite egg-laying in schistosome-infected SCID mice. Nature 356: 604607.
- BARTEL DP. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281297.
- BECKMANN S, BURO C, DISSOUS C, HIRZMANN J AND GREVELDING CG. 2010. The Syk kinase SmTK4 of Schistosoma mansoni is involved in the regulation of spermatogenesis and oogenesis. PLoS Pathog 6: e1000769.
- BEGUN DJ ET AL. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol 5: e310.
- BERRIMAN M ET AL. 2009. The genome of the blood fluke Schistosoma mansoni Nature 460: 352358.
- BIRNEY E ET AL. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799816.
- BOYLE JP, WU XJ, SHOEMAKER CB AND YOSHINO TP. 2003. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol Biochem Parasitol 128: 205215.
- BRITO GC, FACHEL AA, VETTORE AL, VIGNAL GM, GIMBA ER, CAMPOS FS, BARCINSKI MA, VERJOVSKI-ALMEIDA S AND REIS EM. 2008. Identification of protein-coding and intronic noncoding RNAs downregulated in clear cell renal carcinoma. Mol Carcinog 47: 757767.
- BROSNAN CA AND VOINNET O. 2009. The long and the short of noncoding RNAs. Curr Opin Cell Biol 21: 416425.
- CARNINCI P ET AL. 2003. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res 13: 12731289.
- CAWLEY S ET AL. 2004. Unbiased Mapping of Transcription Factor Binding Sites along Human Chromosomes 21 and 22 Points toWidespread Regulation of Noncoding RNAs. Cell 116: 499509.
- CHALMERS IW, MCARDLE AJ, COULSON RM, WAGNER MA, SCHMID R, HIRAI H AND HOFFMANN KF. 2008. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC genomics 9: 89.
- CHEN D, FARWELL MA AND ZHANG B. 2010a. MicroRNA as a new player in the cell cycle. J Cell Physiol 225: 296301.
- CHEN J, YANG Y, GUO S, PENG J, LIU Z, LI J, LIN J AND CHENG G. 2010b. Molecular cloning and expression profiles of Argonaute proteins in Schistosoma japonicum. Parasitol Res 107: 889899.
- CHEN L-L AND CARMICHAEL GG. 2010. Long noncoding RNAs in mammalian cells: what, where, and why? WIREs RNA 1: 19.
- CHENG GF, LIN JJ, SHI Y, JIN YX, FU ZQ, JIN YM, ZHOU YC AND CAI YM. 2005. Dose-dependent inhibition of gynecophoral canal protein gene expression in vitro in the schistosome (Schistosoma japonicum) by RNA interference. Acta Biochim Biophys Sin (Shanghai) 37: 386390.
- CHURCH DM ET AL. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112.
- CHURCHILL GA. 2004. Using ANOVA to analyze microarray data. Biotechniques 37: 173175, 177.
- CLAVERIE JM. 2005. Fewer genes, more noncoding RNA. Science 309: 15291530.
- COPELAND CS, MARZ M, ROSE D, HERTEL J, BRINDLEY PJ, SANTANA CB, KEHR S, ATTOLINI CS AND STADLER PF. 2009. Homology-based annotation of noncoding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum BMC Genomics 10: 464.
- CORE LJ, WATERFALL JJ AND LIS JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 18451848.
- CORRENTI JM, BRINDLEY PJ AND PEARCE EJ. 2005. Long-term suppression of cathepsin B levels by RNA interference retards schistosome growth. Mol Biochem Parasitol 143: 209215.
- COSTA FF. 2010. Non-coding RNAs: Meet thy masters. Bioessays 32: 599608.
- DAHARY D, ELROY-STEIN O AND SOREK R. 2005. Naturally occurring antisense: transcriptional leakage or real overlap? Genome Res 15: 364368.
- DAVIES SJ, GROGAN JL, BLANK RB, LIM KC, LOCKSLEY RM AND MCKERROW JH. 2001. Modulation of blood fluke development in the liver by hepatic CD4+ lymphocytes. Science 294: 13581361.
- DE LUCIA F AND DEAN C. 2010. Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol 14: 168173.
- DE MENDONCA RL, ESCRIVA H, BOUTON D, LAUDET V AND PIERCE RJ. 2000. Hormones and nuclear receptors in schistosome development. Parasitol Today 16: 233240.
- DELCROIX M, SAJID M, CAFFREY CR, LIM KC, DVORAK J, HSIEH I, BAHGAT M, DISSOUS C AND MCKERROW JH. 2006. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J Biol Chem 281: 3931639329.
- DEMARCO R, MATHIESON W, MANUEL SJ, DILLON GP, CURWEN RS, ASHTON PD, IVENS AC, BERRIMAN M, VERJOVSKI-ALMEIDA S AND WILSON RA. 2010. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res 20: 11121121.
- DEMARCO R, OLIVEIRA KC, VENANCIO TM AND VERJOVSKI- ALMEIDA S. 2006. Gender biased differential alternative splicing patterns of the transcriptional cofactor CA150 gene in Schistosoma mansoni Mol Biochem Parasitol 150: 123131.
- DILLON GP, FELTWELL T, SKELTON JP, ASHTON PD, COULSON PS, QUAIL MA, NIKOLAIDOU-KATSARIDOU N, WILSON RA AND IVENS AC. 2006. Microarray analysis identifies genes preferentially expressed in the lung schistosomulum of Schistosoma mansoni Int J Parasitol 36: 18.
- DINDOT SV, PERSON R, STRIVENS M, GARCIA R AND BEAUDET AL. 2009. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res 19: 13741383.
- DINGER ME, AMARAL PP, MERCER TR AND MATTICK JS. 2009. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic 8: 407423.
- DINGUIRARD N AND YOSHINO TP. 2006. Potential role of a CD36-like class B scavenger receptor in the binding of modified low-density lipoprotein (acLDL) to the tegumental surface of Schistosoma mansoni sporocysts. Mol Biochem Parasitol 146: 219230.
- ESCOBEDO G, ROBERTS CW, CARRERO JC AND MORALES- MONTOR J. 2005. Parasite regulation by host hormones: an old mechanism of host exploitation? Trends Parasitol 21: 588593.
- FAGHIRI Z AND SKELLY PJ. 2009. The role of tegumental aquaporin from the human parasitic worm, Schistosoma mansoni, in osmoregulation and drug uptake. FASEB J 23: 27802789.
- FERBEYRE G, SMITH JM AND CEDERGREN R. 1998. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol 18: 38803888.
- FITZPATRICK JM, PEAK E, PERALLY S, CHALMERS IW, BARRETT J, YOSHINO TP, IVENS AC AND HOFFMANN KF. 2009. Anti-schistosomal intervention targets identified by lifecycle transcriptomic analyses. PLoS Negl Trop Dis 3: e543.
- FRANCO GR, ADAMS MD, SOARES MB, SIMPSON AJ, VENTER JC AND PENA SD. 1995. Identification of new Schistosoma mansoni genes by the EST strategy using a directional cDNA library. Gene 152: 141147.
- FRANCO GR, VALADAO AF, AZEVEDO V AND RABELO EM. 2000. The Schistosoma gene discovery program: state of the art. Int J Parasitol 30: 453463.
- FREITAS TC, JUNG E AND PEARCE EJ. 2007. TGF-beta signaling controls embryo development in the parasitic flatworm Schistosoma mansoni PLoS Pathog 3: e52.
- GERSTEIN MB, BRUCE C, ROZOWSKY JS, ZHENG D, DU J, KORBEL JO, EMANUELSSON O, ZHANG ZD, WEISSMAN S AND SNYDER M. 2007. What is a gene, post-ENCODE? History and updated definition. Genome Res 17: 669681.
- GERSTEIN MB ET AL. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775-1787.
- GOBERT GN, MOERTEL L, BRINDLEY PJ AND MCMANUS DP. 2009. Developmental gene expression profiles of the human pathogen Schistosoma japonicum BMC Genomics 10: 128.
- GOLDEN DE, GERBASI VR AND SONTHEIMER EJ. 2008. An inside job for siRNAs. Mol Cell 31: 309-312.
- GOMES MS, CABRAL FJ, JANNOTTI-PASSOS LK, CARVALHO O, RODRIGUES V, BABA EH AND SA RG. 2009. Preliminary analysis of miRNA pathway in Schistosoma mansoni Parasitol Int 58: 61-68.
- GRAVELEY BR ET AL. 2010. The developmental transcriptome of Drosophila melanogaster Nature 471: 473-479.
- GRYSEELS B, POLMAN K, CLERINX J AND KESTENS L. 2006. Human schistosomiasis. Lancet 368: 1106-1118.
- GUTTMAN M ET AL. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223-227.
- HAN ZG, BRINDLEY PJ, WANG SY AND CHEN Z. 2009. Schistosoma genomics: new perspectives on schistosome biology and host-parasite interaction. Annu Rev Genomics Hum Genet 10: 211-240.
- HAO L, CAI P, JIANG N, WANG H AND CHEN Q. 2010. Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum BMC Genomics 11: 55.
- HUANG J, HAO P, CHEN H, HU W, YAN Q, LIU F AND HAN ZG. 2009. Genome-wide identification of Schistosoma japonicum microRNAs using a deep-sequencing approach. PLoS One 4: e8206.
- HU W ET AL. 2003. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat Genet 35: 139-147.
- HUTTENHOFER A, SCHATTNER P AND POLACEK N. 2005. Non-coding RNAs: hope or hype? Trends Genet 21: 289-297.
- JOHNSON JM, EDWARDS S, SHOEMAKER D AND SCHADT EE. 2005. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet 21: 93-102.
- JOLLY ER, CHIN CS, MILLER S, BAHGAT MM, LIM KC, DERISI J AND MCKERROW JH. 2007. Gene expression patterns during adaptation of a helminth parasite to different environmental niches. Genome Biol 8: R65.
- KAPRANOV P ET AL. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484-1488.
- KAUL S ET AL. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana Nature 408: 796-815.
- KEELING PJ AND SLAMOVITS CH. 2005. Causes and effects of nuclear genome reduction. Curr Opin Genet Dev 15: 601-608.
- KHAYATH N, VICOGNE J, AHIER A, BENYOUNES A, KONRAD C, TROLET J, VISCOGLIOSI E, BREHM K AND DISSOUS C. 2007. Diversification of the insulin receptor family in the helminth parasite Schistosoma mansoni Febs J 274: 659-676.
- KIYOSAWA H, MISE N, IWASE S, HAYASHIZAKI Y AND ABE K. 2005. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res 15: 463-474.
- KONG L, ZHANG Y, YE ZQ, LIU XQ, ZHAO SQ, WEI L AND GAO G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345-349.
- KRAUTZ-PETERSON G, BHARDWAJ R, FAGHIRI Z, TARARAM CA AND SKELLY PJ. 2010. RNA interference in schistosomes: machinery and methodology. Parasitology 137: 485-495.
- KRAUTZ-PETERSON G, RADWANSKA M, NDEGWA D, SHOEMAKER CB AND SKELLY PJ. 2007. Optimizing gene suppression in schistosomes using RNA interference. Mol Biochem Parasitol 153: 194-202.
- KRAUTZ-PETERSON G AND SKELLY PJ. 2008a. Schistosoma mansoni: the dicer gene and its expression. Exp Parasitol 118: 122-128.
- KRAUTZ-PETERSON G AND SKELLY PJ. 2008b. Schistosome asparaginyl endopeptidase (legumain) is not essential for cathepsin B1 activation in vivo Mol Biochem Parasitol 159: 54-58.
- KUMAGAI T, OSADA Y, OHTA N AND KANAZAWA T. 2009. Peroxiredoxin-1 from Schistosoma japonicum functions as a scavenger against hydrogen peroxide but not nitric oxide. Mol Biochem Parasitol 164: 26-31.
- KUMAR M, MABALIRAJAN U, AGRAWAL A AND GHOSH B. 2010. Proinflammatory role of let-7 miRNAs in experimental asthma? J Biol Chem 285: le19: author reply le20.
- LI N, MUTHUSAMY S, LIANG R, SAROJINI H AND WANG E. 2010. Increased expression of miR-34a and miR-93 in rat liver during aging; and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev 132: 75-85.
- LOURO R, NAKAYA HI, AMARAL PP, FESTA F, SOGAYAR MC, DA SILVA AM, VERJOVSKI-ALMEIDA S AND REIS EM. 2007. Androgen responsive intronic non-coding RNAs. BMC biology 5: 4.
- LOURO R, SMIRNOVA AS AND VERJOVSKI-ALMEIDA S. 2009. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics 93: 291-298.
- LUO R, XUE X, WANG Z, SUN J, ZOU Y AND PAN W. 2010. Analysis and characterization of the genes encoding the Dicer and Argonaute proteins of Schistosoma japonicum Parasit Vectors 3: 90.
- MATTICK JS. 2003. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25: 930-939.
- MATTICK JS. 2004. RNA regulation: a new genetics? Nat Rev Genet 5: 316-323.
- MATTICK JS. 2009. The genetic signatures of noncoding RNAs. PLoS Genet 5: e1000459.
- MERCER TR, DINGER ME AND MATTICK JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155-159.
- MERRICK JM, OSMAN A, TSAI J, QUACKENBUSH J, LOVERDE PT AND LEE NH. 2003. The Schistosoma mansoni gene index: gene discovery and biology by reconstruction and analysis of expressed gene sequences. J Parasitol 89: 261-269.
- MORALES ME, RINALDI G, GOBERT GN, KINES KJ, TORT JF AND BRINDLEY PJ. 2008. RNA interference of Schistosoma mansoni cathepsin D, the apical enzyme of the hemoglobin proteolysis cascade. Mol Biochem Parasitol 157: 160-168.
- MORTAZAVI A, WILLIAMS BA, MCCUE K, SCHAEFFER L AND WOLD B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621-628.
- NAG A AND JACK T. 2010. Sculpting the flower; the role of microRNAs in flower development. Curr Top Dev Biol 91: 349-378.
- NAKAYA HI, AMARAL PP, LOURO R, LOPES A, FACHEL AA, MOREIRA YB, EL-JUNDI TA, DA SILVA AM, REIS EM AND VERJOVSKI-ALMEIDA S. 2007. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol 8: R43.
- NANA-SINKAM SP, KARSIES T, RISCILI B, EZZIE M AND PIPER M. 2009. Lung microRNA: from development to disease. Expert Rev Respir Med 3: 373-385.
- NDEGWA D, KRAUTZ-PETERSON G AND SKELLY PJ. 2007. Protocols for gene silencing in schistosomes. Exp Parasitol 117: 284-291.
- OLIVEIRA KC, CARVALHO ML, VENANCIO TM, MIYASATO PA, KAWANO T, DEMARCO R AND VERJOVSKIALMEIDA S. 2009. Identification of the Schistosoma mansoni TNF-alpha receptor gene and the effect of human TNF-alpha on the parasite gene expression profile. PLoS Negl Trop Dis 3: e556.
- OLIVER HF, ORSI RH, PONNALA L, KEICH U, WANG W, SUN Q, CARTINHOUR SW, FILIATRAULT MJ, WIEDMANN M AND BOOR KJ. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10: 641.
- OSMAN A, NILES EG, VERJOVSKI-ALMEIDA S AND LOVERDE PT. 2006. Schistosoma mansoni TGF-beta receptor II: role in host ligand-induced regulation of a schistosome target gene. PLoS Pathog 2: e54.
- PEREIRA TC, PASCOAL VD, MARCHESINI RB, MAIA IG, MAGALHAES LA, ZANOTTI-MAGALHAES EM AND LOPES-CENDES I. 2008. Schistosoma mansoni: evaluation of an RNAi-based treatment targeting HGPRTase gene. Exp Parasitol 118: 619-623.
- PRATT AJ AND MACRAE IJ. 2009. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284: 17897-17901.
- REARICK D, PRAKASH A, MCSWEENY A, SHEPARD SS, FEDOROVA L AND FEDOROV A. 2010. Critical association of ncRNA with introns. Nucleic Acids Res 39: 2357-2366.
- REIS EM, LOURO R, NAKAYA HI AND VERJOVSKI-ALMEIDA S. 2005. As antisense RNA gets intronic. Omics 9: 2-12.
- REIS EM ET AL. 2004. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene 23: 6684-6692.
- RINALDI G, MORALES ME, ALREFAEI YN, CANCELA M, CASTILLO E, DALTON JP, TORT JF AND BRINDLEY PJ. 2009. RNA interference targeting leucine aminopeptidase blocks hatching of Schistosoma mansoni eggs. Mol Biochem Parasitol 167: 118-126.
- RINN JL ET AL. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311-1323.
- ROGER E, GRUNAU C, PIERCE RJ, HIRAI H, GOURBAL B, GALINIER R, EMANS R, CESARI IM, COSSEAU C AND MITTA G. 2008. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata). PLoS Negl Trop Dis 2: e330.
- SCHIER AF. 2007. The maternal-zygotic transition: death and birth of RNAs. Science 316: 406-407.
- SHAFFER JP. 1995. Multiple Hypothesis Testing. Annu Rev Psychol 46: 23.
- SIMOES MC, LEE J, DJIKENG A, CERQUEIRA GC, ZERLOTINI A, DA SILVA-PEREIRA RA, DALBY AR, LOVERDE P, EL-SAYED NM AND OLIVEIRA G. 2011. Identification of Schistosoma mansoni microRNAs. BMC Genomics 12: 47.
- SKELLY PJ, DA'DARA A AND HARN DA. 2003. Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. Int J Parasitol 33: 363-369.
- SMITH EM AND GREGORY TR. 2009. Patterns of genome size diversity in the ray-finned fishes. Hydrobiologia 625: 1-25.
- ST LAURENT G 3RD, FAGHIHI MA AND WAHLESTEDT C. 2009. Non-coding RNA transcripts: sensors of neuronal stress, modulators of synaptic plasticity, and agents of change in the onset of Alzheimer's disease. Neurosci Lett 466: 81-88.
- STEINMANN P, KEISER J, BOS R, TANNER M AND UTZINGER J. 2006. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis 6: 411-425.
- THE_C.elegans_SEQUENCING_CONSORTIUM. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012-2018.
- THE_UNIPROT_CONSORTIUM. 2010. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38: D142-148.
- TRINKLEIN ND, ALDRED SF, HARTMAN SJ, SCHROEDER DI, OTILLAR RP AND MYERS RM. 2004. An abundance of bidirectional promoters in the human genome. Genome Res 14: 62-66.
- TUSHER VG, TIBSHIRANI R AND CHU G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116-5121.
- VERJOVSKI-ALMEIDA S AND DEMARCO R. 2011. Gene structure and splicing in schistosomes. J Proteomics doi: 10.1016/j.jprot.2011.03.022.
- VERJOVSKI-ALMEIDA S, LEITE LC, DIAS-NETO E, MENCK CF AND WILSON RA. 2004. Schistosome transcriptome: insights and perspectives for functional genomics. Trends Parasitol 20: 304-308.
- VERJOVSKI-ALMEIDA S, VENANCIO TM, OLIVEIRA KC, ALMEIDA GT AND DEMARCO R. 2007. Use of a 44k oligoarray to explore the transcriptome of Schistosoma mansoni adult worms. Exp Parasitol 117: 236-245.
- VERJOVSKI-ALMEIDA S ET AL. 2003. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni Nature genetics 35: 148-157.
- VERMEIRE JJ, TAFT AS, HOFFMANN KF, FITZPATRICK JM AND YOSHINO TP. 2006. Schistosoma mansoni: DNA microarray gene expression profiling during the miracidium- to-mother sporocyst transformation. Mol Biochem Parasitol 147: 39-47.
- WANG E, MILLER LD, OHNMACHT GA, LIU ET AND MARINCOLA FM. 2000. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol 18: 457-459.
- WANG Z, XUE X, SUN J, LUO R, XU X, JIANG Y, ZHANG Q AND PAN W. 2010. An "in-depth" description of the small non-coding RNA population of Schistosoma japonicum schistosomulum. PLoS Negl Trop Dis 4: e596.
- WASHIETL S, HOFACKER IL, LUKASSER M, HUTTENHOFER A AND STADLER PF. 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383-1390.
- WEBSTER JP, OLIVIERA G, ROLLINSON D AND GOWER CM. 2010. Schistosome genomes: a wealth of information. Trends Parasitol 26: 103-106.
- WERNER A AND BERDAL A. 2005. Natural antisense transcripts: sound or silence? Physiol Genomics 23: 125-131.
- WERNER A AND SWAN D. 2010. What are natural antisense transcripts good for? Biochem Soc Trans 38: 1144-1149.
- WILHELM BT, MARGUERAT S, WATT S, SCHUBERT F, WOOD V, GOODHEAD I, PENKETT CJ, ROGERS J AND BAHLER J. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239-1243.
- WILUSZ JE, SUNWOO H AND SPECTOR DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494-1504.
- WINSTON WM, MOLODOWITCH C AND HUNTER CP. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456-2459.
- WU W, NILES EG AND LOVERDE PT. 2007. Thyroid hormone receptor orthologues from invertebrate species with emphasis on Schistosoma mansoni BMC Evol Biol 7: 150.
- XUE X, SUN J, ZHANG Q, WANG Z, HUANG Y AND PAN W. 2008. Identification and characterization of novel microRNAs from Schistosoma japonicum PLoS One 3: e4034.
- ZHANG Y, LIU XS, LIU QR AND WEI L. 2006. Genomewide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465-3475.
- ZHAO ZR, LEI L, LIU M, ZHU SC, REN CP, WANG XN AND SHEN JJ. 2008. Schistosoma japonicum: inhibition of Mago nashi gene expression by shRNA-mediated RNA interference. Exp Parasitol 119: 379-384.
- ZHOU Y ET AL. 2009. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 460: 345-351.
- ZOU X, JIN YM, LIU PP, WU QJ, LIU JM AND LIN JJ. 2010. RNAi silencing of calcium-regulated heat-stable protein of 24 kDa in Schistosoma japonicum affects parasite growth. Parasitol Res 108: 567-572.
Correspondence to:
Publication Dates
-
Publication in this collection
03 June 2011 -
Date of issue
June 2011
History
-
Received
05 Feb 2011 -
Accepted
28 Apr 2011