Abstracts
Forty-nine Brazilian Dicyma pulvinata isolates were examined by morphological traits and RFLP, RAPD and AFLP analyses. This fungus is a mycoparasite of Microcyclus ulei, the causal agent of the most devastating rubber (Hevea brasiliensis) disease, known as "South American Leaf Blight" (SALB). These isolates were compared with an Indian isolate from Cercosporidium sp., and a French isolate from Cladosporium fulvum. They were also compared with Dicyma ampullifera from Papua New Guinea. The morphological parameters analyzed confirmed the identification of the Brazilian isolates. The graphic representations of the distance matrices of each molecular marker showed similar results. Dicyma pulvinata isolates from M. ulei were closely related, whereas the reference isolates examined were dispersed. Among the D. pulvinata isolates obtained from M. ulei, a significant pairwise distance was obtained, for all the molecular markers, between the isolates from the areas favorable to the occurrence of SALB (North and Northeast of Brazil) and the region of escape for the disease (Mato Grosso State).
fungi; molecular markers; biological control
Quarenta e nove isolados brasileiros de Dicyma pulvinata foram analisados quanto a características morfológicas, análises de RFLP, RAPD e AFLP. Este fungo é um micoparasita do Microcyclus ulei, o agente causal da mais importante doença da seringueira (Hevea brasiliensis), o mal-das-folhas. Estes isolados foram comparados com isolados indianos obtidos a partir de lesões de Cercosporidium sp., com um isolado proveniente da França obtido a partir de lesões de Cladosporium fulvum e com um isolado de D. ampullifera proveniente de Papua-Nova Guiné. A análise de parâmetros morfológicos confirmou a identificação dos isolados brasileiros. As representações gráficas das matrizes de distância de cada marcador molecular mostraram resultados semelhantes. Os isolados de D. pulvinata obtidos de M. ulei mostraram-se intimamente relacionados. Por outro lado, os isolados - referência, incluídos nestes estudos, mostraram-se bastante dispersos. De acordo com a análise de variância molecular, considerando todos os marcadores, os isolados de D. pulvinata obtidos de M. ulei, apresentaram diferença significativa entre isolados procedentes de áreas de cultivo de seringueira favoráveis à ocorrência do mal-das-folhas (regiões norte e nordeste do Brasil) e aqueles de áreas de escape da doença (Mato Grosso).
ARTIGOS ARTICLES
Molecular characterization of Brazilian Dicyma pulvinata isolates* * Part of the Master Dissertation of the first author. Universidade de Brasília (2001)
Caracterização molecular de isolados brasileiros de Dicyma pulvinata
Eder T. Tavares** ** Capes fellowship ; Myrian S. Tigano; Sueli C. M. Mello; Irene Martins; Celia M. T. Cordeiro
Embrapa Recursos Genéticos e Biotecnologia Brasília, DF, Brazil, e-mail: smello@cenargen.embrapa.br
ABSTRACT
Forty-nine Brazilian Dicyma pulvinata isolates were examined by morphological traits and RFLP, RAPD and AFLP analyses. This fungus is a mycoparasite of Microcyclus ulei, the causal agent of the most devastating rubber (Hevea brasiliensis) disease, known as "South American Leaf Blight" (SALB). These isolates were compared with an Indian isolate from Cercosporidium sp., and a French isolate from Cladosporium fulvum. They were also compared with Dicyma ampullifera from Papua New Guinea. The morphological parameters analyzed confirmed the identification of the Brazilian isolates. The graphic representations of the distance matrices of each molecular marker showed similar results. Dicyma pulvinata isolates from M. ulei were closely related, whereas the reference isolates examined were dispersed. Among the D. pulvinata isolates obtained from M. ulei, a significant pairwise distance was obtained, for all the molecular markers, between the isolates from the areas favorable to the occurrence of SALB (North and Northeast of Brazil) and the region of escape for the disease (Mato Grosso State).
Additional keywords: fungi, molecular markers, biological control.
RESUMO
Quarenta e nove isolados brasileiros de Dicyma pulvinata foram analisados quanto a características morfológicas, análises de RFLP, RAPD e AFLP. Este fungo é um micoparasita do Microcyclus ulei, o agente causal da mais importante doença da seringueira (Hevea brasiliensis), o mal-das-folhas. Estes isolados foram comparados com isolados indianos obtidos a partir de lesões de Cercosporidium sp., com um isolado proveniente da França obtido a partir de lesões de Cladosporium fulvum e com um isolado de D. ampullifera proveniente de Papua-Nova Guiné. A análise de parâmetros morfológicos confirmou a identificação dos isolados brasileiros. As representações gráficas das matrizes de distância de cada marcador molecular mostraram resultados semelhantes. Os isolados de D. pulvinata obtidos de M. ulei mostraram-se intimamente relacionados. Por outro lado, os isolados - referência, incluídos nestes estudos, mostraram-se bastante dispersos. De acordo com a análise de variância molecular, considerando todos os marcadores, os isolados de D. pulvinata obtidos de M. ulei, apresentaram diferença significativa entre isolados procedentes de áreas de cultivo de seringueira favoráveis à ocorrência do mal-das-folhas (regiões norte e nordeste do Brasil) e aqueles de áreas de escape da doença (Mato Grosso).
INTRODUCTION
The hyphomycete fungus Dicyma pulvinata (Berk. & Curt.) Arx [=Hansfordia pulvinata (Berk. & Curt.) Hughes] has been found parasitizing phytopathogenic fungi. Cladosporium fulvum Cooke and Cercosporidium personatum Earle, causal agents of tomato (Lycopersicon esculentum Mill.) leaf mould and late leaf spot of peanut (Arachis hypogaea L.), respectively, are some of the fungi colonized by this hyperparasite (Peresse & Picard, 1980; Mitchell et al., 1987). Therefore, D. pulvinata could be considered a potencial biocontrol agent for plant diseases.
South American Leaf Blight (SALB) of Hevea rubber (Hevea brasiliensis), caused by Microcyclus ulei (P. Henn.) Arx, is identified as one of the world's five most threatening plant diseases and it is still endemic to Central and South America. It was first recorded in 1900 on rubber tree in Brazil (Webster & Baulkwill, 1989). Presently the disease extends from Southern Mexico (180 latitude north) to the state of São Paulo in Brazil (240 latitude south) covering Brazil, Bolivia, Colombia, Peru, Venezuela, Guiana, Trinidad, Tobago, Haiti, Panama, Costa Rica, Nicaragua, Salvador, Honduras, Guatemala and Mexico. This disease has been the main constraint for the development of rubber cultivation in Latin America countries (Gasparotto et al., 1997).
To control the disease, it is recommended that H. brasiliensis be planted in escape areas, characterized by a well-defined dry season. Escape areas occur in Goiás, Mato Grosso do Sul, Pernambuco, Maranhão, Espírito Santo, Rio de Janeiro, Paraná, São Paulo, and Minas Gerais, all are non-traditional areas for rubber tree cultivation. Recent studies, however, have indicated the existence of an ecophysiologic variability among M. ulei isolates. If the information about rapid adaptation of the pathogen to environment is confirmed, the control of this disease will be very difficult (Junqueira & Gasparotto, 1991).
Efforts have been made to control this disease, including the use of D. pulvinata (Bettiol, 1991). This mycoparasite was first reported as colonizing black spots produced by M. ulei in the Amazon Region (Junqueira & Gasparoto (1991). It has subsequently spread to different geographic areas (Mello et al., 1999). At present, little is known about the variability among D. pulvinata isolates obtained from M. ulei lesions. Thus, to better exploit the potential of D. pulvinata as a biocontrol agent, the correct identification and characterization of these isolates is critical to better understand the pathogen-host relationship.
Molecular markers have been used to resolve taxonomic problems and to address questions about the genetic variability of fungi. The objectives of this study were to examine the molecular variation of the D. pulvinata isolates obtained from lesions produced by M. ulei on rubber tree leaves from different regions of Brazil; and to compare these isolates with typical D. pulvinata isolates from two other fungal hosts, as well as with an unrelated D. ampullifera isolate. Genomic DNA of Dicyma spp. was subjected to molecular analyses based on AFLPs (Zabeau & Vos, 1993), RAPDs (Welsh & McClelland, 1990; Williams et al., 1990) and RFLPs.
MATERIALS AND METHODS
Fungal isolation
From late February to late December of 1999, a survey was carried out in several rubber producing areas of Brazil. Leaves with M. ulei lesions colonized by fungi, were placed in a paper bag, inside a plastic bag and transported in a cooler. Spores were harvested from lesions, and plated on potato dextrose agar (PDA). After five days, pure cultures were prepared from single conidia and maintained as stock cultures on PDA slants. The isolated fungi were initially identified based on their conidial morphology and growth characteristics.
Fungal culture and DNA extraction
Monoconidial cultures of 49 D. pulvinata isolates collected from different locations in several rubber production areas of Brazil and two others, originating from India and France, were used in this study (Table 1). One isolate of D. ampullifera (CG 704), obtained from soil in Papua New Guinea, was also included as an outgroup. Cultures were maintained on PDA slants, and stored at 8 ºC in the dark until needed. The mycelia were grown in liquid cultures of spores using Richard medium broth (Tuite, 1978), with shaking at 150 rpm, 26 ºC, for seven days. Mycelium was harvested by filtration through filter paper (Whatman No. 1) and stored at -80 ºC. Genomic DNA was obtained from mycelium ground in liquid nitrogen by using the CTAB extraction method (Rogers & Bendich, 1988).
Morphological analyses
Morphological analyses were conducted with cultures of D. pulvinata grown on Richard medium. Four replicate plates were inoculated with 0.5-cm diameter mycelial plugs cut from the margin of seven-day-old colonies of each isolate. Cultures were incubated at 28 ºC and exposed to a 12-h photoperiod under white light of wavelengths between 350 and 750 nm. Colony aspect and conidial morphology were recorded using 20-day-old cultures. Fifty conidia were examined for each replicate.
IGS region amplification and restriction digestion
The IGS region of ribosomal DNA was amplified by PCR using the primers CNL 12 (CTGAACGCCTCTAA GTCAG) and CNS 1 (GAGACAAGCATATGACTACTG) (Appel & Gordon, 1995). The reactions were conducted in a final volume of 50 µl in a microcentrifuge tube containing: 1x Taq DNA polymerase buffer (GIBCO-BRL), 1.5 mM MgCl2, 200 µM of each dNTP, 0.22 µM of the primer CNL 12, 0.28 µM of the primer CNS 1, 20 ng of genomic DNA and 2.5 units of Taq DNA polymerase (GIBCO-BRL). Amplifications were performed using a PTC-100 thermocycler (MJ Research) programmed for 40 cycles of 1 min at 94 ºC, 45 s at 50 ºC, and 2 min at 72 ºC with a final extension of 5 min at 72 ºC. The reaction mixture (5 µl) was run on 1.5% agarose gels, stained with ethidium bromide and visualized under UV illumination to estimate the size of the amplified IGS fragment based on comparison to size markers. Restriction digestions were carried out with 10 µl of the PCR reaction using the restriction enzymes Alu I, Ava II, Cfo I, Mbo I, and Taq I, according to the manufacturers' directions (GIBCO-BRL). Digested DNA was separated on 2.3% agarose gels, stained with ethidium bromide and visualized under UV light.
Random amplified polymorphic DNA (RAPD)
PCR was performed in 15-µl volumes, using a PTC-100 thermocycler (MJ Research). The amplifications were carried out using 20 ng/reaction of each template DNA and the following reaction mix: 4 mM MgCl2, 0.1 mM of each dNTP, 1 µM primer, 1 U of Taq DNA polymerase (GIBCO-BRL) and 1x Taq DNA polymerase buffer (GIBCO-BRL). The temperature program for the reactions was 94 ºC for 3 min, followed by 40 cycles of 94 ºC for 1 min, 35 ºC for 1 min, 72 ºC for 2.5 min, with a final extension at 72 ºC for 7 min (Tigano-Milani et al., 1995a). Eleven 10-mer random primers were used in this study: OPB-01, OPB-04, OPB-08, OPB-09, OPB-11, OPB-15, OPB-18, OPD-01, OPD-08, OPD-11 and OPD-20 (Operon Technologies Inc.). PCR products were separated on 1.5 % agarose gel, stained with ethidium bromide and visualized under UV light. All amplifications were repeated.
Amplified fragment length polymorphism (AFLP)
Genomic DNA (500 ng) was digested with Eco RI (5 U) and ligated to Eco RI adaptors (50 pmol) in a single reaction at 37 ºC for 16 h using standard molecular biology techniques (Sambrook et al., 1989). Ammonium acetate (3 M, pH5.2) and 95% ethanol were added to the reaction mixture, and incubated at room temperature for 30 min. Anchor-annealed Eco RI fragments were precipitated by centrifugation at 13,000 g for 10 min. The pellet was washed with 70 % ethanol, dried, and resuspended in 50 µl of 0.1x TE buffer (Boucias et al., 2000b). A series of four 19-mer, comprised of the Eco RI adapter sequence plus three 3' selective nucleotides (Boucias et al., 2000a) were used for PCR amplifications. The reactions were carried out in 25 µl final volume containing: 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 ìM of single primer, 3 µl anchor-annealed Eco RI fragment, 1 U Taq DNA polymerase (GIBCO-BRL) and 1x Taq DNA polymerase buffer(GIBCO-BRL). Amplifications were performed using a PTC-100 thermocycler (MJ Research) programmed with an initial step of 1 min at 95 ºC followed by 35 cycles of 1 min at 94 ºC, 1 min at 56 ºC, 2.5 min at 72 ºC and a final extension of 5 min at 72 ºC. Amplification products were run on 2% agarose-synergels (Diversified Biotech), stained with ethidium bromide and visualized under UV light.
Analysis of Data
Results obtained from molecular markers IGS, AFLP, and RAPD analyses were used to construct binary matrices. Each of the isolates had been scored for the presence (1) or absence (0) of fragments. A dissimilarity matrix of genetic distances between isolates was calculated based on the method of Nei and Li (1979). Multidimensional scaling (MDS) model was adjusted for each genetic distance matrix using PROC MDS (SAS, 1999) to generate graphic representations of these matrices. Associations among genetic distances for each marker type were analyzed by the Mantel test (Manly, 1997). This procedure gives the product moment correlation (r) and a statistical test to measure the degree of relatedness between two matrices. The relationship among individuals using the molecular markers was also evaluated according to hierarchical structure, using Arlequin v. 2000 (Schneider et al. 1997), considering Brazilian populations of D. pulvinata from the different regions and individuals within regions. The estimate of the variance components associated with the different hierarchical levels was obtained with the aid of AMOVA (Analysis of Molecular Variance).
RESULTS AND DISCUSSION
For all D. pulvinata isolates the conidial size and other morphological characteristics such as type of conidiogenous cells, conidiophores, colony appearance and mycelial growth rates in the culture corresponded to the description of the species (Hughes, 1951). Colonies of D. pulvinata isolates varied from white to grey/greenish, becoming black. Isolates obtained from M. ulei produced exudates that varied in coloration. Mycelium was immersed and superficial. Conidiophores appeared smooth, dark brown or olivaceous brown, bearing a number of primary branches on the upper part. Conidiogenous cells were on branches, polyblastic, discrete, sympodial and deticulate. D. pulvinata conidia were subspherical, aseptate, with a basal scar, olive brown and solitary (data not shown). The mean conidial size for the D. pulvinata isolates obtained from M. ulei varied from 4.3±0.2 µm (CG 766) to 7.3±0.2 µm (CG 777) in diameter, while the means for non-M. ulei hosts isolates, CG 826 and CG 678, varied between 4.2± 0.2 µm and 3.6± 0.1 µm, respectively.
Like other fungi, the morphological parameters of D. pulvinata do not produce information sufficiently clear for differentiating isolates among the species. In order to determine the genetic variability of D. pulvinata isolates obtained from M. ulei in different Brazilian regions, three genetic methods (RFLP, RAPD and AFLP) were used in this study.
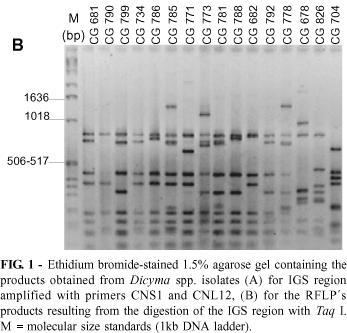
Dicyma pulvinata isolates differed in the size of their IGS region amplified by PCR with primers CNS 1 and CNL 12. The isolates obtained from M. ulei amplified a single double-stranded product of approximately 1.6 kb. However, the isolates collected from different hosts (CG 678 and CG 826) showed a product of approximately 2.0 kb. Although the size of IGS products is the same at the species level for other fungi (Appel & Gordon, 1995; Chillali et al., 1998), polymorphism of the IGS size has already been observed among isolates of Verticillium albo-atrum Reinke & Berthold (Pramateftaki et al., 2000). The isolate of D. ampullifera, CG 704, produced IGS fragments of about 2.6 kb (Figure 1A).
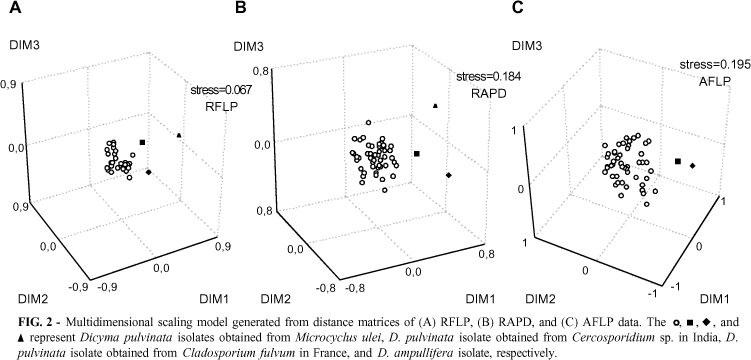
Restriction digestions of IGS revealed RFLPs, reflecting variation in this region of the rDNA among the isolates with the same size of the IGS fragment. The sum of the fragment sizes exceeded the size of the amplified IGS, indicating the existence of different IGS types in the same individual. Each rDNA repeating unit usually consists of a highly conserved region among various organisms and the IGS, which is highly variable in length and in the primary structure among organisms and individuals (Chou & Tsai, 1999). Length variation and sequence polymorphism in IGS have been reported in many fungi (Appel & Gordon, 1996; Kauserud & Schumacher, 2001). From the five enzymes used for the IGS digestion, Taq I produced the most variable patterns (Figure 1B). The RFLP analysis of the IGS region using five restriction enzymes produced 109 scorable characters, from which 107 were polymorphic. Of these selected bands, 42 were due to the inclusion of the two D. pulvinata isolates obtained from non-M. ulei hosts and the outgroup, D. ampullifera (CG 704). The MDS model of the restriction digests of IGS distance matrix data demonstrated the presence of a tight group of D. pulvinata isolates obtained from M. ulei. The two D. pulvinata isolates obtained from non-M. ulei hosts and the outgroup were dispersed from this group (Figure 2A). The digestion with restriction enzyme of this region of the rDNA showed the intraspecific variation among the isolates of D. pulvinata, even among those isolated from the same host in the same country. This method also showed the intraspecific variation within populations of Fusarium oxysporum Smyder & Hansen (Appel & Gordon, 1995).
The IGS region exhibits enough genetic diversity to distinguish intra and interspecies isolates. According to Appel & Gordon (1995), a mutation in an IGS repeat may spread to other spacer regions, leading to the homogenization of the IGS repeats in an individual and in populations that reproduce sexually throughout the parallel evolution mechanism. Thus, the diversity observed among D. pulvinata isolates suggests that this mechanism does not operate, reinforcing the suspicion that this species is an asexual fungus. Sequence data from the IGS region may provide additional information about the phylogeny of D. pulvinata.
The RAPD analysis by using 11 random 10-mer oligonucleotide primers produced a total of 224 scorable polymorphic bands. About 68 % of the bands detected were associated to D. pulvinata species isolated from M. ulei. An example of the patterns obtained with the RAPD analysis is shown in Figure 3. The MDS model for the RAPD data produced a diffuse group of D. pulvinata isolates obtained from M. ulei lesions. Like the RFLP data, D. pulvinata isolates collected from non-M. ulei hosts and the outgroup were also dispersed in this group (Figure 2B). Because of considerable concerns as to the reproducibility of RAPD profiles, all amplifications were repeated and the results were confirmed.
The amplification of the anchor-ligated Eco RI fragments with the four primers produced an array of scorable bands, shown in the example of fingerprintings in Figure 4. A total of 90 scorable characters was selected, all being polymorphic among isolates of Dicyma spp. As observed by RAPD analysis, much of the polymorphism detected with AFLP's was also associated with the D. pulvinata isolates obtained from M. ulei lesions. The MDS model of the AFLP data also produced one dispersed group for the D. pulvinata isolates obtained from M. ulei, while the pulvinata isolates collected from non-M. ulei hosts and the outgroup were dispersed (Figure 2C).
The three molecular markers analyzed proved to be powerful tools for analyzing the intraspecific variability among D. pulvinata isolates. The calculation of the product-moment correlation coefficients for the different combinations of the RFLP, RAPD and AFLP on the Dicyma spp. isolates demonstrated a significant correlation for the combination RFLP and RAPD (r=0.789). The association between RFLP and AFLP or RAPD and AFLP distance matrix produced lower r-values (0.560 and 0.598 respectively).
Spatial distribution of Dicyma spp. isolates based on a combination of genetic distances obtained from AFLP, RAPD and RFLP data sets, showed similar and separate results to the molecular markers. The isolates of D. pulvinata obtained from M. ulei lesions from different regions of Brazil form a group that is distinct from the non-M. ulei isolates and the outgroup (Figure 5). These results suggest a specialization of the isolates to the hosts.
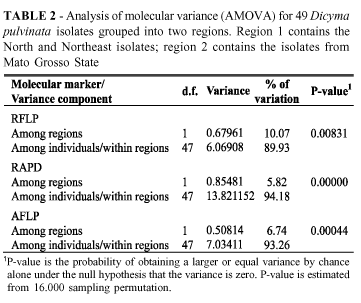
Although the isolates obtained from M. ulei were grouped, the AMOVA tests showed that for each of the three types of molecular markers analyzed, there was a small but significant difference between the 25 isolates obtained from the humid regions, which are favorable to the development of SALB, North and Northeast Brazil, and the 24 isolates from the escape region of the disease, MT, Brazil (Tables 1 and 2). The variance components associated to the variation between these two regions were 0.67961 (P=0.00831), 0.85481 (P< 0.00001) and 0.50814 (P=0.00044) for RFLP, RAPD and AFLP, respectively.
Junqueira et al. (1987) have shown the genetic variability among isolates of the host M. ulei by isozymes analysis, and the correlation between virulence and the isoenzymatic patterns. Considering this, the distance between the two geographical populations of D. pulvinata occurring in lesions of M. ulei should be analyzed taking in consideration the variability of the host.
In spite of the differentiation between the two regions, variation was observed among individuals within regions, indicating the existence of relevant genetic diversity between haplotypes within these populations. This heterogeneity might be important to ensure that the population fits to environment and host variation. In addition, this diversity might be necessary to effectively control the disease by interacting with the parasite, and adapting to changes in host resistance recognition genes. Tigano-Milani et al. (1995b) suggested, for entomopathogenic fungi, that it would be necessary for more than one haplotype to start and maintain epizootics under field conditions. Strongman & MacKay (1993) identified two varieties of the entomopathogenic fungus Hirsutella longicolla Strongman, Eveleigh & Royama, one associated to the first stage of the insect development, and the other associated to the sixth instar of the plague.
The results presented in this study demonstrated the utility of three molecular markers for the characterization of different populations of D. pulvinata.
ACKNOWLEDGEMENTS
Grateful thanks are expressed to William Sihler and Ana Maria C. L. de Barros for technical support; to Bergmann Morais Ribeiro for critical review of the manuscript; to Fundo Nacional do Meio Ambiente (FNMA) and Embrapa for partial support; and to Capes for the award of a scholarship to E. T. Tavares.
LITERATURE CITED
Aceito para publicação em 04/11/2003
Corresponding author: Sueli C. Marques de Mello
- APPEL, D.J. & GORDON, T.R. Intraspecific variation within populations of Fusarium oxysporum based on RFLP analysis of the intergenic spacer region of the rDNA. Experimental Mycology 19:120-128. 1995.
- APPEL, D.J. & GORDON, T.R. Relationship among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Molecular Plant-Microbe Interactions 9:125-138. 1996.
- BETTIOL, W. Seleção de microrganismos antagônicos a fitopatógenos. In: Bettiol, W. (Org.) Controle biológico de doenças de plantas no Brasil. Jaguariúna, SP. EMBRAPA/CNPDA, pp.225-236. 1991.
- BOUCIAS, D.G., STOKES, C., SUAZO, A. & FUNDERBURK, J. AFLP analysis of the entomopathogen Nomuraea rileyi Mycologia 92:638-648. 2000a.
- BOUCIAS, D.G., TIGANO, M.S., SOSA-GOMEZ, D.R., GLARE, T.R. & INGLIS, P.W. Genotypic properties of the entomopathogenic fungus Nomuraea rileyi Biological Control 19:124-138. 2000b.
- CHILLALI, M., IDDER-IGHILI, H., GUILLAUMIN, J.J., MOHAMMED, C., ESCARMANT, B.L. & BOTTON, B. Variation in the ITS and IGS regions of ribosomal DNA among the biological species of European Armillaria Mycological Research 102:533-540. 1998.
- CHOU, C.H. & TSAI, C.C. Genetic variation in the intergenic spacer of ribosomal DNA of Imperata cylindrical (L.) Beauv. Var. major (Cogongrass) populations in Taiwan. Botanical Bulletin of Academia Sinica 40:319-327. 1999.
- GASPAROTTO, L., SANTOS, A.F. dos, PEREIRA, J.C.R. & FERREIRA, F.A. Doenças da seringueira no Brasil. Brasília: Embrapa-SPI: Manaus: Embrapa-CPAA. 1997.
- HUGHES, S.J. Studies on micro-fungi. XI. Mycological Papers 45:1-36. 1951.
- JUNQUEIRA, N.T.V., ALFENAS, A.C., CHAVES, G.M., ZAMBOLIM, L. & GASPAROTTO, L. Variabilidade isoenzimática de isolados de Microcyclus ulei com diferentes níveis de virulência. Fitopatologia Brasileira 12:208-214. 1987.
- JUNQUEIRA, N.T.V. & GASPAROTTO, L. Controle biológico de fungos estromáticos causadores de doenças foliares em seringueira. In: Bettiol, W. (Ed.) Controle biológico de doenças de plantas no Brasil. Jaguariúna: Embrapa-CNPDA, 1991. pp.307-331.
- KAUSERUD, H. & SCHUMACHER, T. Outcrossing or inbreeding: DNA markers provide evidence for type reproductive mode in Phellinus nigrolimitatus (Basidiomycota). Mycological Research 105:676-683. 2001.
- MANLY, B.F.J. Multivariate Statistical Methods: A Primer. 2a ed. Chapman and Hall London. 1997.
- MELLO, S.C.M., SANTOS, M.F., GANGANA, F., KOSOSKI, R.M. & TIGANO, M.S. Levantamento da ocorrência de Dicyma pulvinata parasitando lesões de Microcyclus ulei em seringueira no Brasil. Fitopatologia Brasileira 24:304. 1999. (Resumo)
- MITCHELL, J.K., SMITH, D.H. & TABER, R.A. Potential for biological control of Cercosporidium personatum leafspot of peanuts by Dicyma pulvinata Canadian Journal of Botany 65:2263-2269. 1987.
- NEI, M. & LI, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Science USA 76:5269-5273. 1979.
- PERESSE, M. & PICARD, D.LE. Hansfordia pulvinata, mycoparasite destructeur du Cladosporium fulvum Mycopathologia 71:23-30. 1980.
- PRAMATEFTAKI, P. V., ANTONIOU, P. P. & TYPAS, M.A. The complete DNA sequence of the nuclear ribosomal RNA gene complex of Verticillium dahliae: intraspecific heterogeneity within the intergenic spacer region. Fungal Genetics and Biology 29:19-27. 2000.
- ROGERS, S.O. & BENDICH, A.J. Extraction of DNA from plant tissues. Plant Molecular Biology Manual A6:1-10. 1988.
- SAMBROOK J., FRITSCH E.F. & MANIATIS T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY. 1989.
- SCHNEIDER, S., KUEFFER, J., ROESSLI, B. & EXCOFFIER, L. Arlequin ver. 2000: A software for population genetic data analysis. Genetics and Biometric laboratory, University of Geneva, Switzerland. 1997.
- SAS Institute Inc., SAS/STAT® User's Guide, Version 8, Cary, NC: SAS Institute Inc., 1999.
- STRONGMAN, D.B. & MACKAY, R.M. Discrimination between Hirsutella longicolla var. longicolla and Hirsutella longicolla var. cornuta using random amplified polymorphic DNA fingerprinting. Mycologia 85:65-70. 1993.
- TIGANO-MILANI, M.S., GOMES, A.C.M.M. & SOBRAL, B.W.S. Genetic variability among brazilian isolates of the entomopathogenic fungus Metarhizium anisopliae. Journal of Invertebrate Pathology 65:206-210. 1995a.
- TIGANO-MILANI, M.S., HONEYCUTT, R.J., LACEY, L.A., ASSIS, R., McCLELLAND, M. & SOBRAL, B.W.S. Genetic variability of Paecilomyces fumosoroseus isolates revealed by molecular markers. Journal of Invertebrate Pathology 65:274-282. 1995b.
- TUITE, J. Natural and synthetic culture media: Fungi. In: Reichghl, M. (Ed.) Handbook series in nutrition and food section: Diets, culture media and food supplements. Ohio, CRC Press. 1978.
- WEBSTER, C.C. & BAULKWILL, W.J. Rubber. Longman Group UK Limited, England. 1989.
- WELSH, J. & McCLELLAND, M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research 18:7213-7218. 1990.
- WILLIAMS, J.G.K., KUBELIK, A.R., LIVAK, K.J., RAFALSKI, J.A. & TINGEY, S. DNA polymorphisms amplified by arbitrary primers are useful for genetic markers. Nucleic Acids Research 18:6531-6535. 1990.
- ZABEAU, M. & VOS, P. European Patent Application, publication no: EP0534858. 1993.
Publication Dates
-
Publication in this collection
27 Apr 2004 -
Date of issue
Apr 2004