Abstract
The crosslinking of the Fc fragment (IgG antibody) on a polymer matrix about of dimethylacrylamide (DMA), melamide group (MG) and n-acryloxy succinimide (NAS) was analyzed through Monte Carlo simulation at 277.15K and pH 7, in where Gibs free energy and the dipole moment indicated the spontaneity of the reaction through van der Waals interactions. In addition, the QSAR properties determinated that both the surface area and the volume allow to carry out the physical adsorption of the Fc fragment that was verified through the electronic distribution of the electrostatic potential maps (MESP) where the nucleophilic zones (blue color) and electrophilic (red color) were observed, while the partition coefficient (Log P) indicated the solubility of the process. Subsequently, the analysis of the adsorption of the terbium ion (Tb+3) at 277.15K and a pH 7 in Fc/polymeric matrix was carried out, observing that the Fc fragment presented a flat-on optimization geometry attributed to the Tb+3 that generates electronic repulsions, as well as van der Waals forces, hydrogen bonds derived from the Cys aminoacids formed a polar structure and that was corroborated by the Log P negative. Finally, the surface area and volume determined that Tb+3 adsorption showed an increase in surface area and volume with temperature.
Keywords:
Fc; polymer; QSAR; Gibbs; Monte Carlo
1. Introduction
The development of contrast agents has produced an advance in the analysis of clinical images of biological processes in which the lanthanide elements are analyzed to obtain multimodal images, diagnosis and cancer therapies due to properties of density and magnetic susceptibility[11 Zielhuis, S. W., Nijsen, J. F., Seppenwoolde, J. H., Zonnenberg, B. A., Bakker, C. J., Hennink, W. E., Van Rijk, P. P., & Van Het Schip, A. D. (2005). Lanthanide, bearing microparticulate systems for multi-modality imaging and targeted therapy of cancer. Current Medicinal Chemistry. Anti-Cancer Agents, 5(3), 303-313. http://dx.doi.org/10.2174/1568011053765958. PMid:15992356.
http://dx.doi.org/10.2174/15680110537659...
]. In addition, the lanthanides exhibit a low toxicity attributed to the trivalent state that allows are bonded to ligands that present an oxygen donor, besides are excellent indicators of degenerative diseases due to the antioxidant properties[11 Zielhuis, S. W., Nijsen, J. F., Seppenwoolde, J. H., Zonnenberg, B. A., Bakker, C. J., Hennink, W. E., Van Rijk, P. P., & Van Het Schip, A. D. (2005). Lanthanide, bearing microparticulate systems for multi-modality imaging and targeted therapy of cancer. Current Medicinal Chemistry. Anti-Cancer Agents, 5(3), 303-313. http://dx.doi.org/10.2174/1568011053765958. PMid:15992356.
http://dx.doi.org/10.2174/15680110537659...
,22 Carac, A. (2017). Biological and biomedical applications of the lanthanides compounds: a mini review. Proceedings of the Romanian Academy Series B, 19(2), 69-74. Retrieved in 2020, January 9, from https://pdfs.semanticscholar.org/ad32/2bd248e0bdbf7c2576763353dd72f02ed455.pdf
https://pdfs.semanticscholar.org/ad32/2b...
]. The lanthanide elements have become more important in the identification and treatment of cancer, where the terbium ion (Tb+3) is used in the recognition of protein structures to develop new cancer inhibitors[33 Teo, R. D., Termini, J., & Gray, H. B. (2016). Lanthanides: applications in cancer diagnosis and therapy. Journal of Medicinal Chemistry, 59(13), 6012-6024. http://dx.doi.org/10.1021/acs.jmedchem.5b01975. PMid:26862866.
http://dx.doi.org/10.1021/acs.jmedchem.5...
]. However, the weak adsorption of these ions is attributed to the coordination numbers, the polarity of the transitions f-f and the restriction of rotation.
Nevertheless using polymer matrices the adsorption of lanthanides is carried out and therefore, the design of the controlled release systems are efficient and have no toxicity in the release of peptides, proteins and genes[44 Yoon, M. S., Santra, M., & Ahn, K. H. (2015). Preparation of luminescent lanthanide polymers by ring-opening metathesis polymerization. Tetrahedron Letters, 56(41), 5573-5577. http://dx.doi.org/10.1016/j.tetlet.2015.08.042.
http://dx.doi.org/10.1016/j.tetlet.2015....
,55 Zhao, Z. P., Zheng, K., Li, H. R., Zeng, C. H., Zhong, S., Ng, S. W., Zheng, Y., & Chen, Y. (2018). Structure variation and luminescence of 3d, 2d and 1d lanthanide coordination polymers with 1,3-adamantanediacetic acid. Inorganica Chimica Acta, 482, 340-346. http://dx.doi.org/10.1016/j.ica.2018.06.027.
http://dx.doi.org/10.1016/j.ica.2018.06....
]. There is a diversity of polymers used in the adsorption of biological macromolecules such as polymethyl methacrylate used in the identification of antigen-antibody[66 Lai, X., Gao, G., Watanabe, J., Liu, H., & Shen, H. (2017). Hydrophilic polyelectrolyte multilayers improve the ELISA system: antibody enrichment and blocking free. Polymers, 9(2), 51-63. http://dx.doi.org/10.3390/polym9020051. PMid:30970737.
http://dx.doi.org/10.3390/polym9020051...
], while N-isopropylacrylamide has been studied in the adsorption of immunoglobulin IgG[77 Silva, C. S. O., Baptista, R. P., Santos, A. M., Martinho, J. M. G., Cabral, J. M. S., & Taipa, M. A. (2006). Adsorption of human IgG on to poly(N-isopropylacrylamide)-based polymer particles. Biotechnology Letters, 28(24), 2019-2025. http://dx.doi.org/10.1007/s10529-006-9188-2. PMid:17021661.
http://dx.doi.org/10.1007/s10529-006-918...
]. On the other hand, Polystyrene generates hydrophobic and electrostatic attractions during the immobilization of antibodies in immunoassays[88 Welch, N. G., Scoble, J. A., Muir, B. W., & Pigram, P. J. (2017). Orientation and characterization of immobilized antibodies for improved immunoassays: review. Biointerphases, 12(2), 1-13. http://dx.doi.org/10.1116/1.4978435. PMid:28301944.
http://dx.doi.org/10.1116/1.4978435...
]. Polyacrylamide is the polymer of greatest use in the synthesis of polymer matrices for the adsorption of biological macromolecules in delivery systems due to the characteristics of biocompatibility[11 Zielhuis, S. W., Nijsen, J. F., Seppenwoolde, J. H., Zonnenberg, B. A., Bakker, C. J., Hennink, W. E., Van Rijk, P. P., & Van Het Schip, A. D. (2005). Lanthanide, bearing microparticulate systems for multi-modality imaging and targeted therapy of cancer. Current Medicinal Chemistry. Anti-Cancer Agents, 5(3), 303-313. http://dx.doi.org/10.2174/1568011053765958. PMid:15992356.
http://dx.doi.org/10.2174/15680110537659...
,99 Shmanai, V. V., Nikolayeva, T. A., Vinokurova, L. G., & Litoshka, A. A. (2001). Oriented antibody immobilization to polystyrene macrocarriers for immunoassay modified with hydrazide derivatives of poly(meth)acrylic acid. BMC Biotechnology, 1(1), 4. http://dx.doi.org/10.1186/1472-6750-1-4. PMid:11545680.
http://dx.doi.org/10.1186/1472-6750-1-4...
].
On the other hand, antibodies represent the most important biomolecules in the study of immune systems in the human body for applications in nanobiotechnology and biomedicine[1010 De Michele, C., De Los Rios, P., Foffi, G., & Piazza, F. (2016). Simulation and theory of antibody binding to crowded antigen-covered surfaces. PLoS Computational Biology, 12(3), e1004752. http://dx.doi.org/10.1371/journal.pcbi.1004752. PMid:26967624.
http://dx.doi.org/10.1371/journal.pcbi.1...
11 Hebditch, M., Curtis, R., & Warwicker, J. (2017). Sequence composition predicts immunoglobulin superfamily members that could share the intrinsically disordered properties of antibody ch1 domains. Scientific Reports, 7(1), 12404 . http://dx.doi.org/10.1038/s41598-017-12616-9. PMid:28963509.
http://dx.doi.org/10.1038/s41598-017-126...
-1212 Janeway, C. A., Travers, P., & Walport, M. J. (2001). Immunobiology: the immune system in health and disease. New York: Garland Science.]. Antibodies are constituted by a series of aminoacids; however, performing the computational analysis of the interactions of these molecules with lanthanide elements represents important computational times[1010 De Michele, C., De Los Rios, P., Foffi, G., & Piazza, F. (2016). Simulation and theory of antibody binding to crowded antigen-covered surfaces. PLoS Computational Biology, 12(3), e1004752. http://dx.doi.org/10.1371/journal.pcbi.1004752. PMid:26967624.
http://dx.doi.org/10.1371/journal.pcbi.1...
].
The IgG (Immunoglobulin Gamma) antibody is constituted by glycoproteins of approximately 150 kDa and consists of two heavy chains of 50 kDa and two light chains of 25 kDa, respectively, which are bonded by internal disulfide bonds that have the function of generating the formation of loop to compact the antibody structure[1212 Janeway, C. A., Travers, P., & Walport, M. J. (2001). Immunobiology: the immune system in health and disease. New York: Garland Science.,1313 Hamilton, R. G. (1987). The human IgG subclasses (Doctoral dissertation). Johns Hopkins University, Baltimore, United States.]. The IgG antibody is used in therapies against advanced cancers due to the efficiency of the antigen-binding fragment (Fab) and crystallizable fragment (Fc), which are bonded through a hinge zone that allows the movement of Fab fragments[1212 Janeway, C. A., Travers, P., & Walport, M. J. (2001). Immunobiology: the immune system in health and disease. New York: Garland Science.]. The Fc fragment is composed of the CH2 and CH3 domains that interact with the neonatal Fc receptor (FcRn) of the placenta, liver, mammary glands as well as the intestine in order to control homeostasis and the delivery of IgG to the fetus through of the placenta[1414 Saxena, A., & Wu, D. (2016). Advances in therapeutic Fc engineering-modulation of igg-associated effector functions and serum half-life. Frontiers in Immunology, 7, 580. http://dx.doi.org/10.3389/fimmu.2016.00580. PMid:28018347.
http://dx.doi.org/10.3389/fimmu.2016.005...
15 Gunasekaran, K., Pentony, M., Shen, M., Garrett, L., Forte, C., Woodward, A., Ng, S. B., Born, T., Retter, M., Manchulenko, K., Sweet, H., Foltz, I. N., Wittekind, M., & Yan, W. (2010). Enhacing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. The Journal of Biological Chemistry, 285(25), 19637-19646. http://dx.doi.org/10.1074/jbc.M110.117382. PMid:20400508.
http://dx.doi.org/10.1074/jbc.M110.11738...
16 Zhao, J., Nussinov, R., Wu, W. J., & Ma, B. (2018). In silico methods in antibody design. Antibodies, 7(3), 22-36. http://dx.doi.org/10.3390/antib7030022. PMid:31544874.
http://dx.doi.org/10.3390/antib7030022...
17 Tramontano, A. (2006). The role of molecular modelling in biomedical research. FEBS Letters, 580(12), 2928-2934. http://dx.doi.org/10.1016/j.febslet.2006.04.011. PMid:16647064.
http://dx.doi.org/10.1016/j.febslet.2006...
18 Choe, W., Durgannavar, T. A., & Chung, S. J. (2016). Fc-binding ligands of immunoglobulin g: an overview of high affinity proteins and peptides. Materials, 9(12), 994-1010. http://dx.doi.org/10.3390/ma9120994. PMid:28774114.
http://dx.doi.org/10.3390/ma9120994...
-1919 Lobner, E., Traxlmayr, M. W., Obinger, C., & Hasenhindl, C. (2016). Engineered IgG1-Fc-one fragment to bind them all. Immunological Reviews, 270(1), 113-131. http://dx.doi.org/10.1111/imr.12385. PMid:26864108.
http://dx.doi.org/10.1111/imr.12385...
].
Although there are different computational models for the analysis of antibodies, the force fields of molecular mechanics are more accurate in the calculation of molecular properties mainly in the local reorganization as well as the analysis of the Fc receptor, the CH2 and CH3 domains in addition to the disulfide bonds where the free energy shows information of the molecular interactions and the electronic densities indicate the binding affinity[1616 Zhao, J., Nussinov, R., Wu, W. J., & Ma, B. (2018). In silico methods in antibody design. Antibodies, 7(3), 22-36. http://dx.doi.org/10.3390/antib7030022. PMid:31544874.
http://dx.doi.org/10.3390/antib7030022...
,2020 Hou, T., Chen, K., McLaughlin, W. A., Lu, B., & Wang, W. (2006). Computational analysis and prediction of the binding motif and protein interacting partners of the Abl SH3 domain. PLoS Computational Biology, 2(1), e1. http://dx.doi.org/10.1371/journal.pcbi.0020001. PMid:16446784.
http://dx.doi.org/10.1371/journal.pcbi.0...
].
Computational simulation allows the design and understanding of scaffolding behavior or determination of antibody from QSAR descriptors or properties (Quantitative structure-activity relationship) that allow obtaining information on physicochemical properties, molecular weight, electronic density, surface area, volume, mass and partition coefficient (Log P).
In addition the electrostatic potential maps (MESP) indicate the interactions present in the antibodies[2121 Winkler, J., Armano, G., Dybowski, J. N., Kuhn, O., Ledda, F., & Heider, D. (2011). Computational design of a DNA- and Fc-binding fusion protein. Advances in Bioinformatics, 2011, 457578. http://dx.doi.org/10.1155/2011/457578. PMid:21941539.
http://dx.doi.org/10.1155/2011/457578...
]. Besides, computational simulation allows the design of Fc derivatives for treatments of cancer, viral and immune diseases because they are used as binding media for the selection of functional binders or the obtaining of new proteins for clinical use by calculating the QSAR properties of new biopharmaceuticals[2222 Yang, C., Gao, X., & Gong, R. (2018). Engineering of Fc fragments with optimized physicochemical properties implying improvement of clinical potentials for Fc-based therapeutics. Frontiers in Immunology, 8, 1860. http://dx.doi.org/10.3389/fimmu.2017.01860. PMid:29375551.
http://dx.doi.org/10.3389/fimmu.2017.018...
].
QSAR properties is obtaining using two-stage computational simulations; firstly the polymeric matrix and the Fc fragment and subsequently the adsorption of the lanthanide ion[1010 De Michele, C., De Los Rios, P., Foffi, G., & Piazza, F. (2016). Simulation and theory of antibody binding to crowded antigen-covered surfaces. PLoS Computational Biology, 12(3), e1004752. http://dx.doi.org/10.1371/journal.pcbi.1004752. PMid:26967624.
http://dx.doi.org/10.1371/journal.pcbi.1...
]. These simulations are developed using Monte Carlo modeling to calculate the van der Waals interactions on the surface of the material in order to determine the electrostatic forces when the fragments have strong dipole moments because they predict the orientation on the surface, for example, if there is the influence of the van der Waals forces so the resulting geometry will be flat-on, these characteristics are of vital importance. In the addition of lanthanides to the antibodies in order to increase the efficiency of medical treatment[2323 Castellanos, M. M., Snyder, J. A., Lee, M., Chakravarthy, S., Clark, N. J., Mcauley, A., & Curtis, J. E. (2017). Characterization of monoclonal antibody-protein antigen complexes using small-angle scattering and molecular modeling. Antibodies, 6(4), 25-44. http://dx.doi.org/10.3390/antib6040025. PMid:30364605.
http://dx.doi.org/10.3390/antib6040025...
24 Pellegrini, M., & Doniach, S. (1993). Computer simulation of antibody binding specificity. Proteins, 15(4), 436-444. http://dx.doi.org/10.1002/prot.340150410. PMid:8460113.
http://dx.doi.org/10.1002/prot.340150410...
-2525 Wiseman, M. E., & Frank, C. W. (2012). Antibody adsorption and orientation on hydrophobic surfaces. Langmuir, 28(3), 1765-1774. http://dx.doi.org/10.1021/la203095p. PMid:22181558.
http://dx.doi.org/10.1021/la203095p...
].
Thus, the objective of the research was to use the Monte Carlo modeling to determine the structural properties, the electronic distribution and the Log P of the Tb+3 adsorption on the Fc fragment using a polyacrylamide/melaimide/N-acryloxy succinimide polymeric matrix at 277.15K and a pH of 7.
2. Materials and Methods
Optimization geometry was calculated using the Hyperchem 8v software on a DELL computer with i7 processor. AMBER force field (Assisted Model Building with Energy Refinement) of molecular mechanics was used applying the conjugate gradient method with the Polak-Ribiere algorithm, 19,000 processing cycles, and an RMS (Root mean square) of 0.001 Kcal/(Å-mol) as convergence criteria in order to obtain the minimum of the potential energy surface (PES) according to the Born-Oppenheimer approximation and the Schrodinger equation[2626 Freyhult, E. K., Andersson, K., & Gustafsson, M. G. (2003). Structural modeling extends QSAR analysis of antibody-lysozyme interactions to 3d-qsar. Biophysical Journal, 84(4), 2264-2272. http://dx.doi.org/10.1016/S0006-3495(03)75032-2. PMid:12668435.
http://dx.doi.org/10.1016/S0006-3495(03)...
27 Souza, E. S., Zaramello, L., Kuhnen, C. A., Junkes, B. S., Yunes, R. A., & Heinzen, V. E. F. (2011). Estimating the octanol/water partition coefficient for aliphatic organic compounds using semi-empirical electrotopological index. International Journal of Molecular Sciences, 12(10), 7250-7264. http://dx.doi.org/10.3390/ijms12107250. PMid:22072945.
http://dx.doi.org/10.3390/ijms12107250...
28 Bennour, S., & Louzri, F. (2014). Study of swelling properties and thermal behavior of poly(n,n-dimethylacrylamide-co-maleic acid) based hydrogels. Advances in Chemistry, 2014, 1-10. http://dx.doi.org/10.1155/2014/147398.
http://dx.doi.org/10.1155/2014/147398...
-2929 Fifere, A., Marangoci, N., Maier, S., Coroaba, A., Maftei, D., & Pinteala, M. (2012). Theoretical study on β-cyclodextrin inclusion complexes with propiconazole and protonated propiconazole. Beilstein Journal of Organic Chemistry, 8, 2191-2201. http://dx.doi.org/10.3762/bjoc.8.247. PMid:23365629.
http://dx.doi.org/10.3762/bjoc.8.247...
]. In the design of the polymeric matrix, the geometry was optimized using the model PM3 (Parameterized Model number 3) of quantum mechanics.
Table 1 shows the structural properties of DMA (Dimethylacrylamide), NAS (N acryloxy succinimide) and MG (Melaimide group) respectively, which were used for the design of the polymer matrix of 10 and 40 units. The negative Gibbs free energy shows the thermodynamic stability of the individual molecules[2727 Souza, E. S., Zaramello, L., Kuhnen, C. A., Junkes, B. S., Yunes, R. A., & Heinzen, V. E. F. (2011). Estimating the octanol/water partition coefficient for aliphatic organic compounds using semi-empirical electrotopological index. International Journal of Molecular Sciences, 12(10), 7250-7264. http://dx.doi.org/10.3390/ijms12107250. PMid:22072945.
http://dx.doi.org/10.3390/ijms12107250...
]. On the other hand, the positive Log P determined that the MG is hydrophobic while the negative Log P of the NAS and DMA is slightly hydrophilic, a characteristic that can be used in medical applications as a support in the synthesis of proteins and controlled release systems[2828 Bennour, S., & Louzri, F. (2014). Study of swelling properties and thermal behavior of poly(n,n-dimethylacrylamide-co-maleic acid) based hydrogels. Advances in Chemistry, 2014, 1-10. http://dx.doi.org/10.1155/2014/147398.
http://dx.doi.org/10.1155/2014/147398...
]. The dipole moment indicates that there is a difference in electronegativities attributed to the carbonyl bonds.
Figure 1 shows the mapping of the surface of the isoelectronic density, where the shape and size of the electronic density was appreciated, besides the red coloration represents the negative zone or electrophilic and blue-staining the nucleophilic or positive generated mixture of molecular orbitals[2727 Souza, E. S., Zaramello, L., Kuhnen, C. A., Junkes, B. S., Yunes, R. A., & Heinzen, V. E. F. (2011). Estimating the octanol/water partition coefficient for aliphatic organic compounds using semi-empirical electrotopological index. International Journal of Molecular Sciences, 12(10), 7250-7264. http://dx.doi.org/10.3390/ijms12107250. PMid:22072945.
http://dx.doi.org/10.3390/ijms12107250...
,2929 Fifere, A., Marangoci, N., Maier, S., Coroaba, A., Maftei, D., & Pinteala, M. (2012). Theoretical study on β-cyclodextrin inclusion complexes with propiconazole and protonated propiconazole. Beilstein Journal of Organic Chemistry, 8, 2191-2201. http://dx.doi.org/10.3762/bjoc.8.247. PMid:23365629.
http://dx.doi.org/10.3762/bjoc.8.247...
30 Bivol, V. (2006). Modelling of the 3d-structure of cam:oma photopolymers by using of computational chemistry program. Romanian Journal of Physics, 51(1-2), 269-276. Retrieved in 2020, January 9, from https://pubs.rsc.org/en/content/articlelanding/2016/cp/c5cp03599f#!divAbstract
https://pubs.rsc.org/en/content/articlel...
31 Holstein, P., Harris, R. K., & Say, B. J. (1997). Solid-state 19F NMR investigation of poly(vinylidene fluoride) with high-power proton decoupling. Solid State Nuclear Magnetic Resonance, 8(4), 201-206. http://dx.doi.org/10.1016/S0926-2040(97)00014-3. PMid:9373900.
http://dx.doi.org/10.1016/S0926-2040(97)...
-3232 Mazri, R., Belaidi, S., Kerassa, A., & Lanez, T. (2014). Conformational analysis, substituent effect and structure activity relationships of 16-membered macrodiolides. International Letters of Chemistry, Physics and Astronomy, 33(2), 146-167. http://dx.doi.org/10.18052/www.scipress.com/ILCPA.33.146.
http://dx.doi.org/10.18052/www.scipress....
]. The sequence design of the Fc fragment was carried out by means the amino acid and nucleic acid database in the Hyperchem software[2727 Souza, E. S., Zaramello, L., Kuhnen, C. A., Junkes, B. S., Yunes, R. A., & Heinzen, V. E. F. (2011). Estimating the octanol/water partition coefficient for aliphatic organic compounds using semi-empirical electrotopological index. International Journal of Molecular Sciences, 12(10), 7250-7264. http://dx.doi.org/10.3390/ijms12107250. PMid:22072945.
http://dx.doi.org/10.3390/ijms12107250...
].
MESP of (a) DMA, (b) NAS and (c) MG obtained through the AMBER/PM3 hybrid model. Where, cyan color: carbon, red color: oxygen, white color: hydrogen and purple color: nitrogen, respectively.
Table 2 shows the structural properties of the Fc fragment of the IgG antibody, where the AMBER force field was first used at 298.15K and then the Monte Carlo simulation was used at 277.15K and a pH of 7. The results show an increase in Gibbs free energy due to a structural rearrangement caused by the covalent interactions of the CH2 domain that generate a loss of thermodynamic stability while the CH3 domain remains constant due to a homo-interaction[1919 Lobner, E., Traxlmayr, M. W., Obinger, C., & Hasenhindl, C. (2016). Engineered IgG1-Fc-one fragment to bind them all. Immunological Reviews, 270(1), 113-131. http://dx.doi.org/10.1111/imr.12385. PMid:26864108.
http://dx.doi.org/10.1111/imr.12385...
,2323 Castellanos, M. M., Snyder, J. A., Lee, M., Chakravarthy, S., Clark, N. J., Mcauley, A., & Curtis, J. E. (2017). Characterization of monoclonal antibody-protein antigen complexes using small-angle scattering and molecular modeling. Antibodies, 6(4), 25-44. http://dx.doi.org/10.3390/antib6040025. PMid:30364605.
http://dx.doi.org/10.3390/antib6040025...
]. According to the dipole moment it was observed that at 298.15K it has a value of 1,729 D due to the electrostatic interactions while at 277.15K the dominant interactions are van der Waals that produces a null dipole. The negative partition coefficient demonstrates that the Fc fragment is soluble in water due to the amount of OH groups present in the structure.
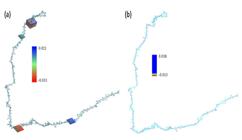
Figure 2 shows the electronic distribution of the Fc fragment at 298.15 and 277.15K and a pH 7 where, the electrophilic zones are observed in red and the nucleophilic zones in blue.
MESP of Fc fragment using Monte Carlo modeling. Where, (a) 298.15 and (b) 277.15K, respectively.
It is also appreciated that the change in temperature increases the positive electronic distribution due to the molecular rearrangement caused by the hydrogen bonds and the van der Waals forces.
3. Results and Discussion
Table 3 shows the structural properties of the ratios 10/1:1 and 40/1:1 of the DMA/(NAS:MG) polymeric matrix calculated at 298K using the AMBER/PM3 hybrid model. It is appreciated that the free energy of Gibbs (ΔG) is spontaneous because the interactions between the molecules generate a rearrangement of charges caused by the heteroatoms, in addition to the difference of the ΔG that can be seen in Tables 1
and 3[2828 Bennour, S., & Louzri, F. (2014). Study of swelling properties and thermal behavior of poly(n,n-dimethylacrylamide-co-maleic acid) based hydrogels. Advances in Chemistry, 2014, 1-10. http://dx.doi.org/10.1155/2014/147398.
http://dx.doi.org/10.1155/2014/147398...
,3333 Chen, J., Jiang, X., Carroll, S., Huang, J., & Wang, J. (2015). Theoretical and experimental investigation of thermodynamics and kinetics of thiol-michael addition reactions: a case study of reversible fluorescent probes for glutathione imaging in single cells. Organic Letters, 17(24), 5978-5981. http://dx.doi.org/10.1021/acs.orglett.5b02910. PMid:26606171.
http://dx.doi.org/10.1021/acs.orglett.5b...
]. The addition of the MG to the polymer generated the Michael reaction (nucleophilic) in the double bond of the structure of the melamide allows a reaction with the functional groups of DMA and NAS attributed to the electron loss of the carbonyls[3434 Hulubei, C. (2008). Functional maleimide-based structural polymers. Revue Roumaine de Chimie, 53(9), 743-752. Retrieved in 2020, January 9, from http://revroum.lew.ro/wp-content/uploads/2008/RRCh_9_2008/Art%2002.pdf
http://revroum.lew.ro/wp-content/uploads...
].
Log P negative determined that the permeation of the hydroxyl group and the amount of carbonyl groups increased the solubility in the polymer chain, being observed in an electronic distribution in red color as shown in Figure 3[3535 Yuan, S., Li, J., Zhu, J., Volodine, A., Li, J., Zhang, G., Van Puyvelde, P., & Van der Bruggen, B. (2018). Hydrophilic nanofiltration membranes with reduced humic acid fouling fabricated from copolymers designed by introducing carboxyl groups in the pendant benzene ring. Journal of Membrane Science, 563, 655-663. http://dx.doi.org/10.1016/j.memsci.2018.06.038.
http://dx.doi.org/10.1016/j.memsci.2018....
].
MESP where (a)10/(1:1) and (b) 40/(1:1) ratios of DMA/(NAS:MG) obtained by the AMBER/PM3 hybrid model.
Table 4 shows the structural properties of the crosslinking of the Fc with the 10 and 40 units of the polymeric matrix applying the Monte Carlo modeling at 277.15K and a pH of 7 were used to control the stability of the melaimide[3636 Marqués-Gallego, P., & De Kroon, A. I. P. M. (2014). Ligation strategies for targeting liposomal nanocarriers. BioMed Research International, 2014, 129458. http://dx.doi.org/10.1155/2014/129458. PMid:25126543.
http://dx.doi.org/10.1155/2014/129458...
]. Gibbs free energy indicated that there is a crosslinking process between Fc and polymer matrix attributed to the nucleophilic and polar character of the Cys (Cysteine) aminoacid that allows acting as a precursor in the formation of disulfide bonds, hydrogen bonds and van der Waals forces present in the Fc fragment on the surface in the polymer matrix producing a molecular rearrangement of the nucleophilic zones first and then the electrophilic zones according to the Michael reaction of thiol groups and maleimides as well as an increase in the surface area that allowed the crosslinking[3737 Maeda, K., Finnie, C., & Svensson, B. (2004). Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. The Biochemical Journal, 378(2), 497-507. http://dx.doi.org/10.1042/bj20031634. PMid:14636158.
http://dx.doi.org/10.1042/bj20031634...
,3838 Zimmermann, J. L., Nicolaus, T., Neuert, G., & Blank, K. (2010). Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nature Protocols, 5(6), 975-985. http://dx.doi.org/10.1038/nprot.2010.49. PMid:20448543.
http://dx.doi.org/10.1038/nprot.2010.49...
].
On the other hand, the aminoacids on Fc are bonded to the polymer by means amino and carboxylic groups producing an amide bond called alpha-aminoacid[3939 Han, G., Chen, S. Y., Gonzalez, V. D., Zunder, E. R., Fantl, W. J., & Nolan, G. P. (2017). Atomic mass tag of bismuth-209 for increasing the immunoassay multiplexing capacity of mass cytometry. Cytometry: Part A, 91(12), 1150-1163. http://dx.doi.org/10.1002/cyto.a.23283. PMid:29205767.
http://dx.doi.org/10.1002/cyto.a.23283...
]. Finally, the sulfhydryl groups of the Cys aminoacids maintain the stability of the Fc fragment avoiding the oxidation process in addition to carrying out the bioconjugation reactions with the melaimide group of the polymer[4040 Chalker, J. M., Bernardes, G. J., Lin, Y. A., & Davis, B. G. (2009). Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chemistry, an Asian Journal, 4(5), 630-640. http://dx.doi.org/10.1002/asia.200800427. PMid:19235822.
http://dx.doi.org/10.1002/asia.200800427...
,4141 Ciborowski, P., & Silberring, J. (2016). In proteomic profiling and analytical chemistry. New York: Elsevier.].
Figure 4 shows that the hydrogen bonding interactions were located in the CH2 domain, however it is appreciated that there is a decrease in the molecular distance that causes an increase in the dipole-dipole, hydrophilic, hydrophobic, and ionic interactions producing a change in the potential surface energy that generates the electrophilic zones in red coloration, while the nucleophilic zones in blue coloration[4242 Ying, T., Ju, T. W., Wang, Y., Prabakaran, P., & Dimitrov, D. S. (2014). Interactions of IgG1 CH2 and CH3 domains with FcRn. Frontiers in Immunology, 5, 146. http://dx.doi.org/10.3389/fimmu.2014.00146. PMid:24765095.
http://dx.doi.org/10.3389/fimmu.2014.001...
,4343 Singh, S. N., Yadav, S., Shire, S. J., & Kalonia, D. S. (2014). Dipole-dipole interaction in antibody solutions: correlation with viscosity behavior at high concentration. Pharmaceutical Research, 31(9), 2549-2558. http://dx.doi.org/10.1007/s11095-014-1352-0. PMid:24639233.
http://dx.doi.org/10.1007/s11095-014-135...
].
MESP of the Fc fragment using the Monte Carlo modeling where, (a) 10 and (b) 40 units of polymeric matrix (DMA/NAS:MG).
Table 5 shows that the adsorption of the terbium ion (Tb+3) at 277.15K and a pH of 7 presents a Gibbs free energy that indicates a rearrangement due to the change in surface area and volume, attributed to the transfer of intramolecular energy originated by the overlap of the resonance energy of the Tb+3 and the energy of the triplet of the aminoacids presents in the Fc fragment[4444 Krepper, W., Satzer, P., Beyer, B. M., & Jungbauer, A. (2018). Temperature dependence of antibody adsorption in protein A affinity chromatography. Journal of Chromatography. A, 1551, 59-68. http://dx.doi.org/10.1016/j.chroma.2018.03.059. PMid:29625770.
http://dx.doi.org/10.1016/j.chroma.2018....
,4545 Arquilla, M., Thompson, L. M., Pearlman, L. F., & Simpkins, H. (1983). Effect of platinum antitumor agents on DMA and MA investigated by terbium fluorescence. Cancer Research, 43(3), 1211-1216. PMid:6186371.].
In addition, the presence of covalent interactions corresponding to Cys aminoacids that allow crosslinking with Tb+3[4646 Vázquez-Ibar, J. L., Weinglass, A. B., & Kaback, H. R. (2002). Engineering a terbium-binding site into an integral membrane protein for luminescence energy transfer. Proceedings of the National Academy of Sciences of the United States of America, 99(6), 3487-3492. http://dx.doi.org/10.1073/pnas.052703599. PMid:11891311.
http://dx.doi.org/10.1073/pnas.052703599...
] as well as van der Waals forces, hydrogen bonds and ionic bonds. On the other hand, the melaimide group in the polymeric matrix allows carrying out the adsorption of thiols by the Cys aminoacids due to the formation of a thioether bond[4747 Ravi, S., Krishnamurthy, V. R., Caves, J. M., Haller, C. A., & Chaikof, E. L. (2012). Maleimide-thiol coupling of a bioactive peptide to an elastin-like protein polymer. Acta Biomaterialia, 8(2), 627-635. http://dx.doi.org/10.1016/j.actbio.2011.10.027. PMid:22061108.
http://dx.doi.org/10.1016/j.actbio.2011....
,4848 Nanda, J. S., & Lorsch, J. R. (2014). Laboratory Methods in Enzymology: protein. Part A: methods in enzymology. London: Elsevier.].
Log P indicates the hydrophilic character attributed to the Cys aminoacid due to the sulfhydryl group and the hydroxyls present in the side chain of polar aminoacids such as Tyr (Tyrosine), Trp (Tryptophan) and Gln (Glutamine)[4848 Nanda, J. S., & Lorsch, J. R. (2014). Laboratory Methods in Enzymology: protein. Part A: methods in enzymology. London: Elsevier.
49 Bulaj, G., Kortemme, T., & Goldenberg, D. P. (1998). Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry, 37(25), 8965-8972. http://dx.doi.org/10.1021/bi973101r. PMid:9636038.
http://dx.doi.org/10.1021/bi973101r...
-5050 Kogan, S., Zeng, Q., Ash, N., & Greenes, R. A. (2001). Problems and challenges in patient information retrieval: a descriptive study. Proceedings - AMIA Symposium, 2001, 329-333. PMid:11825205.]. The control of the temperature at 277.15K is carried out to avoid a total deployment of the CH2 domains that would generate a decrease in the adsorption of Tb+3 as well as a degradation of the polymeric matrix and conformational changes of the Fc fragment[4444 Krepper, W., Satzer, P., Beyer, B. M., & Jungbauer, A. (2018). Temperature dependence of antibody adsorption in protein A affinity chromatography. Journal of Chromatography. A, 1551, 59-68. http://dx.doi.org/10.1016/j.chroma.2018.03.059. PMid:29625770.
http://dx.doi.org/10.1016/j.chroma.2018....
,5151 Ionescu, R. M., Vlasak, J., Price, C., & Kirchmeier, M. (2008). Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. Journal of Pharmaceutical Sciences, 97(4), 1414-1426. http://dx.doi.org/10.1002/jps.21104. PMid:17721938.
http://dx.doi.org/10.1002/jps.21104...
,5252 Hess, B., & Van der Vegt, N. F. A. (2006). Hydration thermodynamic properties of amino acid analogues: a systematic comparison of biomolecular force fields and water models. The Journal of Physical Chemistry B, 110(35), 17616-17626. http://dx.doi.org/10.1021/jp0641029. PMid:16942107.
http://dx.doi.org/10.1021/jp0641029...
].
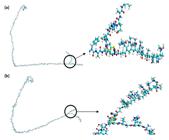
Figure 5 shows that the interactions with the Tb+3 ion produce α-helix and β-sheet structure of the Fc fragment attributed to the increase in structural stability that maintains the conformation[2323 Castellanos, M. M., Snyder, J. A., Lee, M., Chakravarthy, S., Clark, N. J., Mcauley, A., & Curtis, J. E. (2017). Characterization of monoclonal antibody-protein antigen complexes using small-angle scattering and molecular modeling. Antibodies, 6(4), 25-44. http://dx.doi.org/10.3390/antib6040025. PMid:30364605.
http://dx.doi.org/10.3390/antib6040025...
,3838 Zimmermann, J. L., Nicolaus, T., Neuert, G., & Blank, K. (2010). Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nature Protocols, 5(6), 975-985. http://dx.doi.org/10.1038/nprot.2010.49. PMid:20448543.
http://dx.doi.org/10.1038/nprot.2010.49...
]. In addition, the electron density distribution indicates the presence of an electronic repulsion due to the polarization effected of the Tb+3 ion obtaining a geometry of the Fc fragment of the flat-on type[2525 Wiseman, M. E., & Frank, C. W. (2012). Antibody adsorption and orientation on hydrophobic surfaces. Langmuir, 28(3), 1765-1774. http://dx.doi.org/10.1021/la203095p. PMid:22181558.
http://dx.doi.org/10.1021/la203095p...
,5353 Ning, L., Zhang, L., Hu, L., Yang, F., Duan, C., & Zhang, Y. (2011). DFT calculations of crystal-field parameters for the lanthanide ions in the LaCl3 crystal. Journal of Physics Condensed Matter, 23(20), 205502. http://dx.doi.org/10.1088/0953-8984/23/20/205502. PMid:21540498.
http://dx.doi.org/10.1088/0953-8984/23/2...
]. On the other hand, the carbonyl groups form bonds with the Tb+3 due to the transfer of electrons to the amino groups attributed to the delocalized electrons[5454 Rzączyńska, Z., Woźniak, M., Wołodkiewicz, W., Ostasz, A., & Pikus, S. (2007). Thermal properties of lanthanide(III) complexes with 5-amino-1,3-benzenedicarboxylic acid. Journal of Thermal Analysis and Calorimetry, 88(3), 871-876. http://dx.doi.org/10.1007/s10973-005-7463-4.
http://dx.doi.org/10.1007/s10973-005-746...
,5555 Beck, A., Goetsch, L., Dumontet, C., & Corvaïa, N. (2017). Strategies and challenges for the next generation of antibody-drug conjugates. Nature Reviews. Drug Discovery, 16(5), 315-337. http://dx.doi.org/10.1038/nrd.2016.268. PMid:28303026.
http://dx.doi.org/10.1038/nrd.2016.268...
]. Furthermore, pH control at 7 is carried out because at acid levels the aminoacid chain are protonated, that is, an electrostatic repulsion is generated, thus producing the denaturation of Fc[5656 Kanmert, D. (2011). Structure and interactions of human IgG-Fc (Thesis dissertation). Linkoping University, Linkoping, Sweden.].
MESP of the adsorption of the Tb+3 using the Monte Carlo modeling where (a) 10 and (b) 40 units of polymeric matrix (DMA/NAS:MG). Terbium ion is denoted in black color.
4. Conclusions
The Monte Carlo simulation allowed to analyze first the crosslinking between the polymer matrix and the crystallizable fragment (Fc) and then the adsorption of the terbium ion (Tb+3) at 277.15K and a pH of 7, observing that the Gibbs free energies indicated a spontaneous process of the adsorption of the terbium ion using the Cys aminoacid.
In addition, the partition coefficient (Log P) determined that there is solubility in the adsorption attributed to the sulfhydryl and hydroxyl groups present in the Fc and through the melaimide group it was established that there is greater adsorption of Fc in the polymeric matrix. Conformational surface changes were determined due to the van der Waals forces that originated a flat-on structure of the Fc as well as hydrophilic and hydrophobic interactions and hydrogen bonds that were verified by changes in the electronic distribution of the MESPs where the increase of the nucleophilic zones and subsequently the electrophilic zones is appreciated as established in the Michael reaction.
5. Acknowledgements
I would like to say, thanks so much at Dr. Edgar Arriaga from School of Chemistry-University of Minnesota (Twin Cities) for all the support during the stay.
-
How to cite: Rangel Vázquez, N. A. (2020). Adsorption of terbium ion on Fc/dymethylacrylamide: application of Monte Carlo simulation. Polímeros: Ciência e Tecnologia, 30(1), e2020006. https://doi.org/10.1590/0104-1428.08419
6. References
-
1Zielhuis, S. W., Nijsen, J. F., Seppenwoolde, J. H., Zonnenberg, B. A., Bakker, C. J., Hennink, W. E., Van Rijk, P. P., & Van Het Schip, A. D. (2005). Lanthanide, bearing microparticulate systems for multi-modality imaging and targeted therapy of cancer. Current Medicinal Chemistry. Anti-Cancer Agents, 5(3), 303-313. http://dx.doi.org/10.2174/1568011053765958 PMid:15992356.
» http://dx.doi.org/10.2174/1568011053765958 -
2Carac, A. (2017). Biological and biomedical applications of the lanthanides compounds: a mini review. Proceedings of the Romanian Academy Series B, 19(2), 69-74. Retrieved in 2020, January 9, from https://pdfs.semanticscholar.org/ad32/2bd248e0bdbf7c2576763353dd72f02ed455.pdf
» https://pdfs.semanticscholar.org/ad32/2bd248e0bdbf7c2576763353dd72f02ed455.pdf -
3Teo, R. D., Termini, J., & Gray, H. B. (2016). Lanthanides: applications in cancer diagnosis and therapy. Journal of Medicinal Chemistry, 59(13), 6012-6024. http://dx.doi.org/10.1021/acs.jmedchem.5b01975 PMid:26862866.
» http://dx.doi.org/10.1021/acs.jmedchem.5b01975 -
4Yoon, M. S., Santra, M., & Ahn, K. H. (2015). Preparation of luminescent lanthanide polymers by ring-opening metathesis polymerization. Tetrahedron Letters, 56(41), 5573-5577. http://dx.doi.org/10.1016/j.tetlet.2015.08.042
» http://dx.doi.org/10.1016/j.tetlet.2015.08.042 -
5Zhao, Z. P., Zheng, K., Li, H. R., Zeng, C. H., Zhong, S., Ng, S. W., Zheng, Y., & Chen, Y. (2018). Structure variation and luminescence of 3d, 2d and 1d lanthanide coordination polymers with 1,3-adamantanediacetic acid. Inorganica Chimica Acta, 482, 340-346. http://dx.doi.org/10.1016/j.ica.2018.06.027
» http://dx.doi.org/10.1016/j.ica.2018.06.027 -
6Lai, X., Gao, G., Watanabe, J., Liu, H., & Shen, H. (2017). Hydrophilic polyelectrolyte multilayers improve the ELISA system: antibody enrichment and blocking free. Polymers, 9(2), 51-63. http://dx.doi.org/10.3390/polym9020051 PMid:30970737.
» http://dx.doi.org/10.3390/polym9020051 -
7Silva, C. S. O., Baptista, R. P., Santos, A. M., Martinho, J. M. G., Cabral, J. M. S., & Taipa, M. A. (2006). Adsorption of human IgG on to poly(N-isopropylacrylamide)-based polymer particles. Biotechnology Letters, 28(24), 2019-2025. http://dx.doi.org/10.1007/s10529-006-9188-2 PMid:17021661.
» http://dx.doi.org/10.1007/s10529-006-9188-2 -
8Welch, N. G., Scoble, J. A., Muir, B. W., & Pigram, P. J. (2017). Orientation and characterization of immobilized antibodies for improved immunoassays: review. Biointerphases, 12(2), 1-13. http://dx.doi.org/10.1116/1.4978435 PMid:28301944.
» http://dx.doi.org/10.1116/1.4978435 -
9Shmanai, V. V., Nikolayeva, T. A., Vinokurova, L. G., & Litoshka, A. A. (2001). Oriented antibody immobilization to polystyrene macrocarriers for immunoassay modified with hydrazide derivatives of poly(meth)acrylic acid. BMC Biotechnology, 1(1), 4. http://dx.doi.org/10.1186/1472-6750-1-4 PMid:11545680.
» http://dx.doi.org/10.1186/1472-6750-1-4 -
10De Michele, C., De Los Rios, P., Foffi, G., & Piazza, F. (2016). Simulation and theory of antibody binding to crowded antigen-covered surfaces. PLoS Computational Biology, 12(3), e1004752. http://dx.doi.org/10.1371/journal.pcbi.1004752 PMid:26967624.
» http://dx.doi.org/10.1371/journal.pcbi.1004752 -
11Hebditch, M., Curtis, R., & Warwicker, J. (2017). Sequence composition predicts immunoglobulin superfamily members that could share the intrinsically disordered properties of antibody ch1 domains. Scientific Reports, 7(1), 12404 . http://dx.doi.org/10.1038/s41598-017-12616-9 PMid:28963509.
» http://dx.doi.org/10.1038/s41598-017-12616-9 -
12Janeway, C. A., Travers, P., & Walport, M. J. (2001). Immunobiology: the immune system in health and disease New York: Garland Science.
-
13Hamilton, R. G. (1987). The human IgG subclasses (Doctoral dissertation). Johns Hopkins University, Baltimore, United States.
-
14Saxena, A., & Wu, D. (2016). Advances in therapeutic Fc engineering-modulation of igg-associated effector functions and serum half-life. Frontiers in Immunology, 7, 580. http://dx.doi.org/10.3389/fimmu.2016.00580 PMid:28018347.
» http://dx.doi.org/10.3389/fimmu.2016.00580 -
15Gunasekaran, K., Pentony, M., Shen, M., Garrett, L., Forte, C., Woodward, A., Ng, S. B., Born, T., Retter, M., Manchulenko, K., Sweet, H., Foltz, I. N., Wittekind, M., & Yan, W. (2010). Enhacing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. The Journal of Biological Chemistry, 285(25), 19637-19646. http://dx.doi.org/10.1074/jbc.M110.117382 PMid:20400508.
» http://dx.doi.org/10.1074/jbc.M110.117382 -
16Zhao, J., Nussinov, R., Wu, W. J., & Ma, B. (2018). In silico methods in antibody design. Antibodies, 7(3), 22-36. http://dx.doi.org/10.3390/antib7030022 PMid:31544874.
» http://dx.doi.org/10.3390/antib7030022 -
17Tramontano, A. (2006). The role of molecular modelling in biomedical research. FEBS Letters, 580(12), 2928-2934. http://dx.doi.org/10.1016/j.febslet.2006.04.011 PMid:16647064.
» http://dx.doi.org/10.1016/j.febslet.2006.04.011 -
18Choe, W., Durgannavar, T. A., & Chung, S. J. (2016). Fc-binding ligands of immunoglobulin g: an overview of high affinity proteins and peptides. Materials, 9(12), 994-1010. http://dx.doi.org/10.3390/ma9120994 PMid:28774114.
» http://dx.doi.org/10.3390/ma9120994 -
19Lobner, E., Traxlmayr, M. W., Obinger, C., & Hasenhindl, C. (2016). Engineered IgG1-Fc-one fragment to bind them all. Immunological Reviews, 270(1), 113-131. http://dx.doi.org/10.1111/imr.12385 PMid:26864108.
» http://dx.doi.org/10.1111/imr.12385 -
20Hou, T., Chen, K., McLaughlin, W. A., Lu, B., & Wang, W. (2006). Computational analysis and prediction of the binding motif and protein interacting partners of the Abl SH3 domain. PLoS Computational Biology, 2(1), e1. http://dx.doi.org/10.1371/journal.pcbi.0020001 PMid:16446784.
» http://dx.doi.org/10.1371/journal.pcbi.0020001 -
21Winkler, J., Armano, G., Dybowski, J. N., Kuhn, O., Ledda, F., & Heider, D. (2011). Computational design of a DNA- and Fc-binding fusion protein. Advances in Bioinformatics, 2011, 457578. http://dx.doi.org/10.1155/2011/457578 PMid:21941539.
» http://dx.doi.org/10.1155/2011/457578 -
22Yang, C., Gao, X., & Gong, R. (2018). Engineering of Fc fragments with optimized physicochemical properties implying improvement of clinical potentials for Fc-based therapeutics. Frontiers in Immunology, 8, 1860. http://dx.doi.org/10.3389/fimmu.2017.01860 PMid:29375551.
» http://dx.doi.org/10.3389/fimmu.2017.01860 -
23Castellanos, M. M., Snyder, J. A., Lee, M., Chakravarthy, S., Clark, N. J., Mcauley, A., & Curtis, J. E. (2017). Characterization of monoclonal antibody-protein antigen complexes using small-angle scattering and molecular modeling. Antibodies, 6(4), 25-44. http://dx.doi.org/10.3390/antib6040025 PMid:30364605.
» http://dx.doi.org/10.3390/antib6040025 -
24Pellegrini, M., & Doniach, S. (1993). Computer simulation of antibody binding specificity. Proteins, 15(4), 436-444. http://dx.doi.org/10.1002/prot.340150410 PMid:8460113.
» http://dx.doi.org/10.1002/prot.340150410 -
25Wiseman, M. E., & Frank, C. W. (2012). Antibody adsorption and orientation on hydrophobic surfaces. Langmuir, 28(3), 1765-1774. http://dx.doi.org/10.1021/la203095p PMid:22181558.
» http://dx.doi.org/10.1021/la203095p -
26Freyhult, E. K., Andersson, K., & Gustafsson, M. G. (2003). Structural modeling extends QSAR analysis of antibody-lysozyme interactions to 3d-qsar. Biophysical Journal, 84(4), 2264-2272. http://dx.doi.org/10.1016/S0006-3495(03)75032-2 PMid:12668435.
» http://dx.doi.org/10.1016/S0006-3495(03)75032-2 -
27Souza, E. S., Zaramello, L., Kuhnen, C. A., Junkes, B. S., Yunes, R. A., & Heinzen, V. E. F. (2011). Estimating the octanol/water partition coefficient for aliphatic organic compounds using semi-empirical electrotopological index. International Journal of Molecular Sciences, 12(10), 7250-7264. http://dx.doi.org/10.3390/ijms12107250 PMid:22072945.
» http://dx.doi.org/10.3390/ijms12107250 -
28Bennour, S., & Louzri, F. (2014). Study of swelling properties and thermal behavior of poly(n,n-dimethylacrylamide-co-maleic acid) based hydrogels. Advances in Chemistry, 2014, 1-10. http://dx.doi.org/10.1155/2014/147398
» http://dx.doi.org/10.1155/2014/147398 -
29Fifere, A., Marangoci, N., Maier, S., Coroaba, A., Maftei, D., & Pinteala, M. (2012). Theoretical study on β-cyclodextrin inclusion complexes with propiconazole and protonated propiconazole. Beilstein Journal of Organic Chemistry, 8, 2191-2201. http://dx.doi.org/10.3762/bjoc.8.247 PMid:23365629.
» http://dx.doi.org/10.3762/bjoc.8.247 -
30Bivol, V. (2006). Modelling of the 3d-structure of cam:oma photopolymers by using of computational chemistry program. Romanian Journal of Physics, 51(1-2), 269-276. Retrieved in 2020, January 9, from https://pubs.rsc.org/en/content/articlelanding/2016/cp/c5cp03599f#!divAbstract
» https://pubs.rsc.org/en/content/articlelanding/2016/cp/c5cp03599f#!divAbstract -
31Holstein, P., Harris, R. K., & Say, B. J. (1997). Solid-state 19F NMR investigation of poly(vinylidene fluoride) with high-power proton decoupling. Solid State Nuclear Magnetic Resonance, 8(4), 201-206. http://dx.doi.org/10.1016/S0926-2040(97)00014-3 PMid:9373900.
» http://dx.doi.org/10.1016/S0926-2040(97)00014-3 -
32Mazri, R., Belaidi, S., Kerassa, A., & Lanez, T. (2014). Conformational analysis, substituent effect and structure activity relationships of 16-membered macrodiolides. International Letters of Chemistry, Physics and Astronomy, 33(2), 146-167. http://dx.doi.org/10.18052/www.scipress.com/ILCPA.33.146
» http://dx.doi.org/10.18052/www.scipress.com/ILCPA.33.146 -
33Chen, J., Jiang, X., Carroll, S., Huang, J., & Wang, J. (2015). Theoretical and experimental investigation of thermodynamics and kinetics of thiol-michael addition reactions: a case study of reversible fluorescent probes for glutathione imaging in single cells. Organic Letters, 17(24), 5978-5981. http://dx.doi.org/10.1021/acs.orglett.5b02910 PMid:26606171.
» http://dx.doi.org/10.1021/acs.orglett.5b02910 -
34Hulubei, C. (2008). Functional maleimide-based structural polymers. Revue Roumaine de Chimie, 53(9), 743-752. Retrieved in 2020, January 9, from http://revroum.lew.ro/wp-content/uploads/2008/RRCh_9_2008/Art%2002.pdf
» http://revroum.lew.ro/wp-content/uploads/2008/RRCh_9_2008/Art%2002.pdf -
35Yuan, S., Li, J., Zhu, J., Volodine, A., Li, J., Zhang, G., Van Puyvelde, P., & Van der Bruggen, B. (2018). Hydrophilic nanofiltration membranes with reduced humic acid fouling fabricated from copolymers designed by introducing carboxyl groups in the pendant benzene ring. Journal of Membrane Science, 563, 655-663. http://dx.doi.org/10.1016/j.memsci.2018.06.038
» http://dx.doi.org/10.1016/j.memsci.2018.06.038 -
36Marqués-Gallego, P., & De Kroon, A. I. P. M. (2014). Ligation strategies for targeting liposomal nanocarriers. BioMed Research International, 2014, 129458. http://dx.doi.org/10.1155/2014/129458 PMid:25126543.
» http://dx.doi.org/10.1155/2014/129458 -
37Maeda, K., Finnie, C., & Svensson, B. (2004). Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. The Biochemical Journal, 378(2), 497-507. http://dx.doi.org/10.1042/bj20031634 PMid:14636158.
» http://dx.doi.org/10.1042/bj20031634 -
38Zimmermann, J. L., Nicolaus, T., Neuert, G., & Blank, K. (2010). Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nature Protocols, 5(6), 975-985. http://dx.doi.org/10.1038/nprot.2010.49 PMid:20448543.
» http://dx.doi.org/10.1038/nprot.2010.49 -
39Han, G., Chen, S. Y., Gonzalez, V. D., Zunder, E. R., Fantl, W. J., & Nolan, G. P. (2017). Atomic mass tag of bismuth-209 for increasing the immunoassay multiplexing capacity of mass cytometry. Cytometry: Part A, 91(12), 1150-1163. http://dx.doi.org/10.1002/cyto.a.23283 PMid:29205767.
» http://dx.doi.org/10.1002/cyto.a.23283 -
40Chalker, J. M., Bernardes, G. J., Lin, Y. A., & Davis, B. G. (2009). Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chemistry, an Asian Journal, 4(5), 630-640. http://dx.doi.org/10.1002/asia.200800427 PMid:19235822.
» http://dx.doi.org/10.1002/asia.200800427 -
41Ciborowski, P., & Silberring, J. (2016). In proteomic profiling and analytical chemistry New York: Elsevier.
-
42Ying, T., Ju, T. W., Wang, Y., Prabakaran, P., & Dimitrov, D. S. (2014). Interactions of IgG1 CH2 and CH3 domains with FcRn. Frontiers in Immunology, 5, 146. http://dx.doi.org/10.3389/fimmu.2014.00146 PMid:24765095.
» http://dx.doi.org/10.3389/fimmu.2014.00146 -
43Singh, S. N., Yadav, S., Shire, S. J., & Kalonia, D. S. (2014). Dipole-dipole interaction in antibody solutions: correlation with viscosity behavior at high concentration. Pharmaceutical Research, 31(9), 2549-2558. http://dx.doi.org/10.1007/s11095-014-1352-0 PMid:24639233.
» http://dx.doi.org/10.1007/s11095-014-1352-0 -
44Krepper, W., Satzer, P., Beyer, B. M., & Jungbauer, A. (2018). Temperature dependence of antibody adsorption in protein A affinity chromatography. Journal of Chromatography. A, 1551, 59-68. http://dx.doi.org/10.1016/j.chroma.2018.03.059 PMid:29625770.
» http://dx.doi.org/10.1016/j.chroma.2018.03.059 -
45Arquilla, M., Thompson, L. M., Pearlman, L. F., & Simpkins, H. (1983). Effect of platinum antitumor agents on DMA and MA investigated by terbium fluorescence. Cancer Research, 43(3), 1211-1216. PMid:6186371.
-
46Vázquez-Ibar, J. L., Weinglass, A. B., & Kaback, H. R. (2002). Engineering a terbium-binding site into an integral membrane protein for luminescence energy transfer. Proceedings of the National Academy of Sciences of the United States of America, 99(6), 3487-3492. http://dx.doi.org/10.1073/pnas.052703599 PMid:11891311.
» http://dx.doi.org/10.1073/pnas.052703599 -
47Ravi, S., Krishnamurthy, V. R., Caves, J. M., Haller, C. A., & Chaikof, E. L. (2012). Maleimide-thiol coupling of a bioactive peptide to an elastin-like protein polymer. Acta Biomaterialia, 8(2), 627-635. http://dx.doi.org/10.1016/j.actbio.2011.10.027 PMid:22061108.
» http://dx.doi.org/10.1016/j.actbio.2011.10.027 -
48Nanda, J. S., & Lorsch, J. R. (2014). Laboratory Methods in Enzymology: protein. Part A: methods in enzymology London: Elsevier.
-
49Bulaj, G., Kortemme, T., & Goldenberg, D. P. (1998). Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry, 37(25), 8965-8972. http://dx.doi.org/10.1021/bi973101r PMid:9636038.
» http://dx.doi.org/10.1021/bi973101r -
50Kogan, S., Zeng, Q., Ash, N., & Greenes, R. A. (2001). Problems and challenges in patient information retrieval: a descriptive study. Proceedings - AMIA Symposium, 2001, 329-333. PMid:11825205.
-
51Ionescu, R. M., Vlasak, J., Price, C., & Kirchmeier, M. (2008). Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. Journal of Pharmaceutical Sciences, 97(4), 1414-1426. http://dx.doi.org/10.1002/jps.21104 PMid:17721938.
» http://dx.doi.org/10.1002/jps.21104 -
52Hess, B., & Van der Vegt, N. F. A. (2006). Hydration thermodynamic properties of amino acid analogues: a systematic comparison of biomolecular force fields and water models. The Journal of Physical Chemistry B, 110(35), 17616-17626. http://dx.doi.org/10.1021/jp0641029 PMid:16942107.
» http://dx.doi.org/10.1021/jp0641029 -
53Ning, L., Zhang, L., Hu, L., Yang, F., Duan, C., & Zhang, Y. (2011). DFT calculations of crystal-field parameters for the lanthanide ions in the LaCl3 crystal. Journal of Physics Condensed Matter, 23(20), 205502. http://dx.doi.org/10.1088/0953-8984/23/20/205502 PMid:21540498.
» http://dx.doi.org/10.1088/0953-8984/23/20/205502 -
54Rzączyńska, Z., Woźniak, M., Wołodkiewicz, W., Ostasz, A., & Pikus, S. (2007). Thermal properties of lanthanide(III) complexes with 5-amino-1,3-benzenedicarboxylic acid. Journal of Thermal Analysis and Calorimetry, 88(3), 871-876. http://dx.doi.org/10.1007/s10973-005-7463-4
» http://dx.doi.org/10.1007/s10973-005-7463-4 -
55Beck, A., Goetsch, L., Dumontet, C., & Corvaïa, N. (2017). Strategies and challenges for the next generation of antibody-drug conjugates. Nature Reviews. Drug Discovery, 16(5), 315-337. http://dx.doi.org/10.1038/nrd.2016.268 PMid:28303026.
» http://dx.doi.org/10.1038/nrd.2016.268 -
56Kanmert, D. (2011). Structure and interactions of human IgG-Fc (Thesis dissertation). Linkoping University, Linkoping, Sweden.
Publication Dates
-
Publication in this collection
01 June 2020 -
Date of issue
2020
History
-
Received
09 Jan 2020 -
Reviewed
27 Mar 2020 -
Accepted
14 Apr 2020