Abstract
Nosocomial infections (NI) result in considerably high mortality and morbidity rates, especially among pediatric patients. Considering current worldwide changes, information about the occurrence of pathogens and susceptibility tests are now seen as decisive for optimizing treatment. The purpose of this research was to determine the frequency of microorganisms, antimicrobial and genetic profiles, and risk factors associated with nosocomial infections in a teaching hospital in Campo Grande, Mato Grosso do Sul. From January 1998 to December 1999, 108 patients were characterized as having nosocomial infection, from which 137 pathogens were isolated. Identification and antimicrobial susceptibility was determined by conventional and automated techniques. Staphylococcus aureus and Klebsiella pneumoniae strains were characterized by Pulsed Field Gel Electrophoresis (PFGE). Pathogens were most often isolated from infants one-month old or younger, and bloodstream infections were the most frequent. The main isolated agents isolated were: coagulase-negative staphylococci (38), Pseudomonas aeruginosa (19), S. aureus (26), K. pneumoniae (18), and Candida spp. (13). The risk conditions that were most closely related to NI acquisition were: prolonged hospital stays (69.4%), prematurity (60.9%) and exposure to high-risk device procedures (95.4%). Ciprofloxacin and imipenem were the most effective drugs, inhibiting all or almost all of the Enterobacteriaceae, P. aeruginosa and Acinetobacter calcoaceticus isolates. Only 23% of the S. aureus samples were resistant to oxacillin. Genomic typing revealed 10 distinct patterns for S. aureus and 13 for K. pneumoniae, suggesting that most them did not belong to the same clone. PFGE was effective in differentiating the strains.
Infant; nosocomial infection; antimicrobial susceptibility
Surveillance of pediatric infections in a teaching hospital in Mato Grosso do Sul, Brazil
Chang M.R.; Carvalho N.C.P.; Oliveira A.L.L.; Moncada P.M.F.; Moraes B.A.; Asensi M.D.
Federal University of Mato Grosso do Sul, Campo Grande/MS; Department of Bacteriology IOC/F FIOCRUZ, Rio de Janeiro/RJ, Brazil
Correspondence Correspondence to Dr. Marilene Rodrigues Chang Rua Uricuri, 608. Vila Olinda, Campo Grande MS, Brasil Zip code: 79060-040 E-mail: mchang@nin.ufms.br
ABSTRACT
Nosocomial infections (NI) result in considerably high mortality and morbidity rates, especially among pediatric patients. Considering current worldwide changes, information about the occurrence of pathogens and susceptibility tests are now seen as decisive for optimizing treatment. The purpose of this research was to determine the frequency of microorganisms, antimicrobial and genetic profiles, and risk factors associated with nosocomial infections in a teaching hospital in Campo Grande, Mato Grosso do Sul. From January 1998 to December 1999, 108 patients were characterized as having nosocomial infection, from which 137 pathogens were isolated. Identification and antimicrobial susceptibility was determined by conventional and automated techniques. Staphylococcus aureus and Klebsiella pneumoniae strains were characterized by Pulsed Field Gel Electrophoresis (PFGE). Pathogens were most often isolated from infants one-month old or younger, and bloodstream infections were the most frequent. The main isolated agents isolated were: coagulase-negative staphylococci (38), Pseudomonas aeruginosa (19), S. aureus (26), K. pneumoniae (18), and Candida spp. (13). The risk conditions that were most closely related to NI acquisition were: prolonged hospital stays (69.4%), prematurity (60.9%) and exposure to high-risk device procedures (95.4%). Ciprofloxacin and imipenem were the most effective drugs, inhibiting all or almost all of the Enterobacteriaceae, P. aeruginosa and Acinetobacter calcoaceticus isolates. Only 23% of the S. aureus samples were resistant to oxacillin. Genomic typing revealed 10 distinct patterns for S. aureus and 13 for K. pneumoniae, suggesting that most them did not belong to the same clone. PFGE was effective in differentiating the strains.
Key Words: Infant, nosocomial infection, antimicrobial susceptibility.
Nosocomial infections result in considerable mortality and morbidity, especially among pediatric patients [1-3]. In national and international research, special attention has been given to infant bacteremia, due to the risk factors to which they are exposed during their treatment and maintenance [1,4-7].
Considering ongoing worldwide changes, vigilance of predominant pathogens and susceptibility tests are now seen as decisive for optimizing treatment [7-9]. Surveillance data allow hospitals to take appropriate intervention measures to control the infection [5].
Phenotypic and genotypic methods have been used in epidemiologic studies. Recent reports consider them to be effective and useful tools for elucidating the means of dissemination and infection reservoirs [3,7-11].
We determined the frequency of microorganisms and the risk factors associated with nosocomial infections in a teaching hospital in Campo Grande, Mato Grosso do Sul and examined whether the bacteria involved in nosocomial infections at the pediatric units had the same antimicrobial and genetic profile as other bacteria isolated at the same unit.
Materials and Methods
Description of Hospital and Population. Bacterial strains were isolated from patients attended at pediatric units of the UFMS teaching hospital (Campo Grande-MS, Brazil). This hospital provides assistance to patients of all ages, from various parts of the city of Campo Grande in the State of Mato Grosso do Sul. From January 1998 to December 1999, 108 patients were characterized as having NI according to the Center for Disease Control (CDC) criteria [12]; in which 137 pathogens were isolated from these patients.
The data collected for each infection included the date, the site of infection and demographic characteristics of the patients.
Microorganisms, Identification and Susceptibility Testing. The culture and identification were made using the Vitek automated system [13]. Antimicrobial susceptibility was determined by the disk diffusion method, according to National Committee for Clinical Laboratory Standards recommendations NCCLS, M100-S9 [14] for the drugs: CRO- ceftriaxone (30mg), TAZ- ceftazidime (30mg), TAX- cefotaxime (30mg) CLO chloramphenicol (30mg) and TET- tetracycline (30mg). The MIC values (mg/mL) were determined with the Vitek System for AMP- ampicillin, CFL- cephalothin, TAZ- ceftazidime, TAX- cefotaxime, CPE- cefepime, FOX- cefoxitin, AZT- aztreonam; IMP- imipenem, TOB- tobramycin, GEN- gentamicin, AMI- amikacin CIP- ciprofloxacin, TSX trimethoprin-sulfametoxazole, OXA oxacillin and VAN - vancomycin.
Quality control was carried out using standard strains of E. coli (ATCC 25922), P. aeruginosa (ATCC 27953) and S. aureus (ATCC 25923). For presumptive extended-spectrum b lactamase production we used the screening tests for ESBL K. pneumoniae recommended by NCCLS M100-S9: ceftazidime zone <0.22 mm, cefotaxime zone <0.27 mm and ceftriaxone zone <0.25 mm. ESBL production was determined by using E-test strips (AB Biodisk, Sona, SW). If MIC TZ/MIC TZL < 8, ESBL was considered negative and if the MIC of TZ/MIC TZL > 8, ESBL was considered positive.
Pulsed Field Gel Electrophoresis PFGE. Chromosomal DNA analysis of S. aureus (17) and K. pneumoniae (19) were performed by PFGE, according to procedures described by Sader et al., 1994 [8] and Weller et al., 1997 [9], respectively. DNA restriction was processed using Sma I for S. aureus and Xba I for K. pneumoniae. The fragments were separated using the CHEF DRIII pulsed field electrophoresis system (BioRad, Laboratories, Richmond, Calif.). The fragments were stained with ethidium bromide and photographed. All agarose gels were prepared in duplicate.
Band patterns were analyzed using Gel Compar II (Applied Maths, Belgium). The relation among fingerprints was determined by constructing a similarity matrix using Dice's coefficient with 1.5% position tolerance; a dendrogram was generated using the UPGMA clustering algorithm. The definition of the clone structure of isolates was made according to Tenover, 1997 [10].
Epidemiological Analysis of Patients. An epidemiological record with demographics and microbiological data was prepared for each NI case. Data were organized with Excel 7.0 software (Microsoft) and analyzed with Epi Info (version 6.04: CDC, Atlanta, USA)
Statistical Analysis. A univariate analysis was performed to determine significance in comparisons between groups. X2 and Fisher tests were also used, where appropriate.
Results
Several parameters of the epidemiological data were analyzed. The general features of the 108 pediatric patients and the 59 controls group can be observed in Table 1. The average age of the patients was 4 months and 21 days for the NI group. Fifty-one percent of the infants were male. The pathogens were most often isolated from one-month- old or younger newborns. In this age range, bloodstream infections were the most frequent. The patients that had NI stayed in the hospital for up to about 28 days. Of the 108 children, 20 (18.5%) died during hospitalization.
Microorganisms were most frequently isolated from blood samples (85), followed by catheter tips (23), wound secretions (11), eyes, ears or nose secretions (3) and others (3). Overall, 66 cases of bloodstream infection (BSI) were recorded (48.2%), 22 cases of pneumonia (PNE, 16.1%), 2 lower respiratory tract infections, excluding pneumonia (LRTI, 1.5 %), 14 eye, ear and nose infections (EENI, 10.2%), 9 systemic infections (Sis, 6.6%), 8 gastrointestinal system infections (GSI, 5.8%), 6 cases of skin and soft tissue infection (SSTI, 6.44%), 7 surgical wound infections (SWI, 5.1%), 2 urinary tract infections (UTI, 1.5%) and 1 case of central nervous system infection (CNSI, 1.7%). The distribution of the site of the infection varied in the different pediatric units; BSI, pneumonia and GSI were most frequent among newborns kept in the neonatal intensive-care unit (NICU), while skin and soft tissue infection were most common in patients kept in infirmaries (Table 2).
The risk conditions that were mostly related to NI acquisition were prolonged hospitalization (75, 69.4%), prematurity (44, 60.9%) and exposure to high-risk devices and procedures (103, 95.4%) (Table 1). The most frequently isolated pathogens were coagulase-negative staphylococci (CoNS) (38), P. aeruginosa (19), S. aureus (26), K. pneumoniae (18), and Candida spp. (13) (Table 3). Death occurred in 7 (10.9%) of the 64 Gram-positive cocci (GPC), 14 (23%) of 60 Gram-negative bacilli (GNB), and 2 (15. %) of the 13 fungal infection patients.
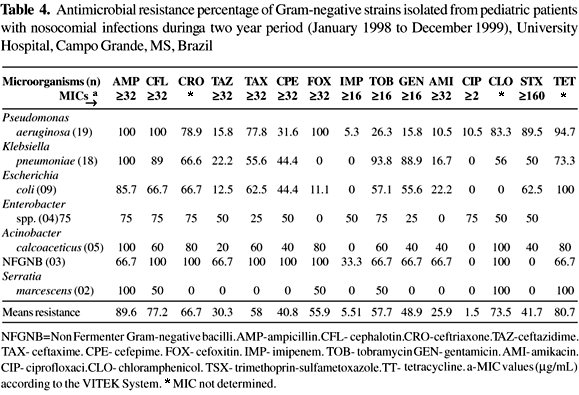
The resistance of GNB to the antimicrobials was determined (Table 4). Ciprofloxacin and imipenem were the most effective drugs, inhibiting all, or almost all, of the Enterobacteriaceae, P. aeruginosa and A. calcoaceticus isolates; resistance to ampicillin was more frequent (89.6%, MIC> 32 mg/mL). All of the P. aeruginosa, K. pneumoniae and Serratia isolates were resistant to this antibiotic.
The frequency of resistance of the P. aeruginosa isolates to tobramycin was 26.3% (MIC >16mg/mL), followed by 15.8% to gentamicin (MIC >16mg/mL) and 10.5% to amikacin (MIC >32mg/mL). The most effective cephalosporin was ceftazidime, as only 15.8% of the isolates were resistant (MIC >32 mg/mL).
Almost ninety percent of the K. pneumoniae isolates were resistant to cephalothin (MIC >32 mg/mL) and resistance to other cephalosporins varied: 66.6% to ceftriaxone, 22.2% to ceftazidime (MIC >32 mg/mL) and 44.4% to cefepime (MIC >32 mg/mL). No strains were resistant to cefoxitin. The distribution of antimicrobial resistance, the frequency of K. pneumoniae producing ESBL and their genetic profile are shown in Table 5.
All E. coli isolates were fully susceptible to imipenem, ciprofloxacin and chloramphenicol. The frequency of resistant to ceftazidime and cefoxitin was 12.5% and 11.1%, respectively. Resistance against the aminoglycosides ranged from 22.2 % to 57.1%, while 65.5% of E. coli strains were resistant to trimethoprin-sulfametoxazole and 100% were resistant to tetracycline.
Fifty percent of the Enterobacter strains were resistant to cefotaxime, cefoxitin, aminoglycosides, trimethoprin-sulfametoxazole and to tetracycline. A high frequency of resistance (75%) was also found against other cephalosporins (cephalotin, ceftriaxone and ceftazidime), chloramphenicol and ampicillin.
The most active compounds against Acinetobacter calcoaceticus (100% susceptibility) were imipenem and ciproloxacin. Among the cephalosporins, ceftazidime gave the lowest percentage of resistance (20%). All isolates were resistant to ampicillin and chloramphenicol, while the frequencies of resistance to tetracycline and tobramycin were 80% and 60%, respectively, in vitro.
All NFGNB isolates were resistant to cephalotin, ceftriaxone, cefotaxime, cefepime and chloramphenicol. Almost sixty-seven percent of these microorganisms were resistant to ceftazidime, ampicillin, aminoglycosides and tetracycline.
The two Serratia isolates were fully susceptible to the third and fourth generation antibiotics, including imipenem, ciprofloxacin, amikacin and gentamicin, and to trimethoprin-sulfametoxazole.
According to the resistance profile of the Staphylococci species, 100% were sensitive to vancomycin (MIC <1m/mL). Twenty-three percent of S. aureus isolates were resistant to oxacillin. The MIC value >4m/mL for oxacillin was considered resistant for all Staphylococci species. All of the S. simulans (3) and S. hominis (2) isolates were resistant to this drug. A high frequency of resistance to oxacilina also observed among S. epidermides (13, 84.6%), S. haemolyticus (7, 71.4%), S. auriculares (2, 50%) and other CoNS (10, 60%) strains.Overall, the mean percentage of CoNS and Staphylococcus species isolates resistant to oxacillin was 66.6% and 61.1%, respectively.
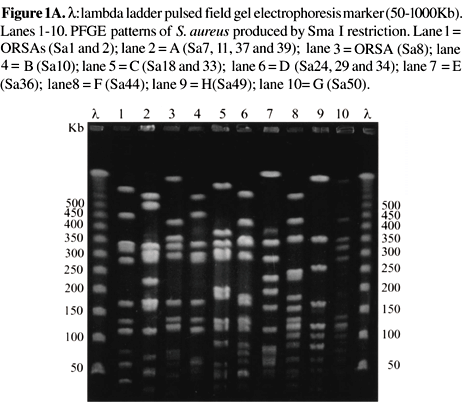
Genomic typing revealed 10 distinct patterns for S. aureus (Figure 1 A). PFGE pattern A was present in 4 strains, isolated from two wound secretions, one ocular secretion and one bloodstream sample. The infections occurred between June/98 and April/99 and the patients were hospitalized in three different units: infirmary, nursery and the neonatal intensive care unit. PFGE pattern D was found in three isolates, responsible for BSI, SWI and skin infection. All the patients were interned in the same infirmary between November 98 and March 99. Clone C of S. aureuswas found in two isolates from newborns treated in the nursery between March and November 99, with systemic and bloodstream infection, respectively. The dendrogram generated by Gel Compar II (Figure 1 B) revealed that 3 ORSA strains belong to two unrelated genetic clones.
We detected 13 different profiles (A M) in the PFGE analysis of 18 K. pneumoniae (Figure 2A). PFGE pattern A was present in 3 isolates, responsible for three BSIs, which occurred between June and July 98 and all these patients were in NICU. PFGE pattern B was found in three isolates, responsible for two BSIs and one ocular infection. One patient with BSI and another with an ocular infection were in the NICU between July and October 98, and the other BSI case occurred in the nursery ward in November 1999. A dendrogram constructed from all profiles showed that the ESBL producing strains with patterns A and H had 80% similarity (Figure 2B).
Discussion
We observed that the frequency of infection was inversely related to the age of the patient; this was quite similar to what was found in some previous studies [1,3]. The duration of hospitalization was longer for children with NI than for the control group (28 versus 13 days).
We found that the frequency of BSI (48.2%) was high, similar to the Raymond study [16], and respiratory diseases were more frequent (16.1%) than previously reported [5].
BSIs were most frequent in newborns (36, 52.2%), and 26 (70.6%) of these subjects had an umbilical catheter. Univariate analysis revealed that invasive procedures, such as catheters, ventilatory support and parenteral nutrition, as well as nasogastric sounds are associated with a risk of nosocomial infections. In addition, prolonged hospitalization may have favored the occurrence of nosocomial infections, as described by other researchers [1-4,17]. In recent years, several reports [4,6] have emphasized that the knowledge of risk factors is useful, so that precautions can be taken to protect patients identified as highly susceptible to infection, to reinforce rational means to administrate and to adequately dose antibiotics, and to avoid invasive procedures that can provoke bacteremia.
The most frequently isolated microorganisms: CoNS, P. aeruginosa, S. aureus, K. pneumoniae and Candida spp. were also described in previous reports as NI agents [2,4,16,18]. Coagulase-negative staphylococci and Candida spp. are emerging as important nosocomial pathogens. We found that P. aeruginosa and K. pneumonia strains were responsible for many BSI cases, and were associated with prematurity (3/7 and 3/5, respectively) and high mortality rates (2/7, and 4/9, respectively).
The results of this in vitro research demonstrate that carbapenem and fluoroquinolone have broad activity against many common aerobic Gram-negative bacteria. These data are in general agreement with other previously reported studies [17,19]. Even though fourth generation chephalosporin is generally effective against Enterobacteriaceae and P. aeruginosa, we found a higher percentage of resistant isolates (29.1%) than the 18.6% observed by Mendes, 2000 [20]. Frequencies of resistance vary greatly among countries, hospital units and even patients [16].
We found cefoxitin, ciprofloxacin and imipenem to be the most effective agents for treatment of K. pneumoniae at the UFMS University Hospital; however these antibiotics are used in our institution only as a last option, since imipenem very quickly induces ESBL production [4] and fluoroquinolone is not yet licensed for use in children, because of its toxicity to cartilage in animal models [21].
ESBL production was detected in 4 K. pneumoniae isolates (23.5%), which is lower than levels found in other Brazilian hospitals [7,20,21]. Despite the small number of strains, this finding is clinically significant because it may result in clinical failure when cephalosporins are used for treatment. The continuous use of ampicillin and gentamicin, which is a policy of our hospital, can lead to selective pressure able to promote resistance to these drugs (Table 4). The K. pneumoniae isolates gave 15 different antimicrobial resistance profiles (ARPs) in which 12 and 15 ARPs were related to a specific genetic profile (A and B, respectively).
Oxacillin-resistant S. aureus is an important agent of NI and it is frequently transmitted in hospital and perinatal units [22,23]. In our study, S. aureus oxacillin-resistance was less frequent (23.1%) than found in other hospitals in Brazil [7,17] and none of these isolates were vancomycin-resistant. The percentage resistance of CoNS against oxacillin was higher (66.6%), similar to results of other researchers [4,7,16],
Genomic typing revealed considerable genetic variety; there were 10 distinct patterns for S. aureus and 13 for K. pneumoniae, suggesting that most did not belong to the same clone, however we observed small outbreaks of both genera in our study.
Different clones of S. aureus identified with the PFGE technique (A, B, E and F) had the same antimicrobial profile; this was also found by other authors [23]. Consequently, an identical profile does not imply that the same clone is involved, as was once thought. Analyzing the clinical data of the patients and the PFGE patterns of the S. aureus strains, little or no relation between infection, locality or period of hospitalization was found.
Genetic similarity was estimated by comparing PFGE patterns. There was a low degree of similarity between the ORSA strains (67-71%), indicating distinct infection sources. The similarity of OSSA isolates varied from 53% to 75%, indicating a high degree of genetic diversity. The two strains of ORSA, which had the same genetic profile, had a 75% similarity with the OSSA of group B.
Klebsiella pneumoniae infections did not represent outbreaks; rather they were a series of sporadic infections caused by different strains. However, there was possibly a small outbreak caused by strains with closely related A patterns in patients in the NICU.
Two ESBL producing K. pneumoniae (Kpne7 and 9) shared the same ARPs and genetic profile (A). Nevertheless, within these same PFGE A patterns, there was one K. pneumoniae isolate that did not produce the ESBL enzyme. These results are in agreement with those of other researchers who reported strains of the same clone that have different ARPs [7,9,11].
We conclude that it is important to screen pediatric patients for NI agents and that the PFGE method is useful for typing pathogens in epidemiological studies.
Acknowledgements
We thank Yeda Bispo, nurse of the infection control service for her support in collecting patients records. We also thank Márcio M. Loureiro and Izabel dos Santos Rodrigues for their technical assistance with PFGE analysis and Frederico Molina Cohr for his assistance with statistical analysis.
Received on 08 March 2002
revised 13 May 2002
- 1. Jarvis W.R. Epidemiology of nosocomial infections in pediatric patients. The Pediatr Infect Dis J 1987;6(4):344-51.
- 2. Moore D.L. Nosocomial infection in newborn nursery and neonate intensive unit. In: Mayhall. C. G.. ed. Hospital epidemiology and infection control.Willian & Wilkins 1996;535-64.
- 3. Villari P., Lacuzi L., Torre L., Scarcella A. Molecular epidemiology as an effective tool in the surveillance of infections in the neonatal intensive care unit. J Infect 1998;37:274-81.
- 4. Cordero L., Sananes M., Leona W. A. Bloodstream infections in a neonatal intensive-care unit: 12 years experience with an antibiotic control program. Infect Control Hosp Epidemiol 1999;20:242 -6.
- 5. Gaynes R. P., Edwards J.R., Jarwia W.R., et al. National Nosocomial Infections Surveillance System. Nosocomial infections among neonates in high-risk nurseries in the United States. Pediatrics 1996;98:357-61.
- 6. Paganini H.R., Rodriguez-Brieschke T., Casimir L., Seu S. Risk factors for nosocomial bacterial infection in children: A case-control study. Medicina 1999;59(1):43-8.
- 7. Sader H.S., Sampaio J.L.M., Zocolli C., Jones R.N. Results of 1997 SENTRY antimicrobial surveillance program in three Brazilian medical centers. BJID 1999;3(2):63-79.
- 8. Sader H.S., Pignatari A.C., Hollis R.J., Jones R.N. Evaluation of interhospital spread of methicillin-resistant Staphylococcus aureus in São Paulo, Brazil, using pulsed field gel electrophoresis of chromosomal DNA. Infect Control Hosp Epidemiol 1994;15(5):320-3.
- 9. Weller T.M.A., Mackenzie F.M., Forbes K.J. Molecular epidemiology of a large outbreak of multiresistant Klebsiella pneumoniae J Med Microbiol 1997;46:921-6.
- 10. Tenover F. C., Arbeit R.D., Goering R.V. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect Control Hosp Epidemiol 1997;18:426-39.
- 11. Loureiro M.M., Moraes B.A., Mendonça V.L.F.D., et al. Molecular epidemiology of extend-spectrum b Lactamase-producing Klebsiella pneumoniae isolated from neonatal intensive care unit patients involved in hospital infection cases in Rio de Janeiro Brazil. Rev Latino Americana de Microbiologia 2001;43:88-95.
- 12. Garner J.S., Jarvis W. R., Emori T.G., et al. CDC definitions for nosocomial infections. Am J Infect Control 1988;15:128-40.
- 13. Arbeit R.D. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P. R., Baron L.J., Pfaller M.A., et al [eds.] Manual of Clinical Microbiology 6th ed ASM Press 1995:190-208.
-
14National Committee for Clinical Laboratory Standards.1999. Performance standards for antimicrobial disk susceptibility testing. Approved standards. NCCLS approved standard M100-S9. National Committee for Clinical Laboratory Standards, Wayne. PA.
- 15. Brodie S.E., Sands K.E., Gray J.E., et al. Occurrence of nosocomial bloodstream infections in six neonatal intensive care units. Pediatr Infect Dis 2000;19(1): 56-62.
- 16. Raymond J., Aujard Y. Nosocomial infections in pediatric patients: a European, multicenter prospective study. Infect Control Hosp Epidemiol 2000;21:260-3.
- 17. De Moraes B.A., Cravo C.A.N., Loureiro M.M., et al. Epidemiological analysis of bacterial strains involved in hospital infection in a university hospital from Brazil. Rev Inst Med Trop 2000;42(4):201-7.
- 18. Oto M.A.L., Samamé MM., Camponovo R.C., et al. Infecciones bacterianas neonatales. Rev Chil Pediatr 1994;65(2):107-13.
- 19. Pfaller M. A., Herwaldt Loreen A. The clinical microbiology laboratory and infection control: emerging pathogens. antimicrobial resistance and new technology. Clin Infect Dis 1997;25:858-70.
- 20. Mendes C., Hsiung A., Kiffer C., et al. Evaluation of the in vitro activity of 9 antimicrobials against bacterial strains isolated from patients in care unit in Brazil: MYSTIC antimicrobial surveillance program. BJID 2000;4:236-44.
- 21. Linder N.R., Dagan J., Kuint N., et al. Ventriculitis caused by Klebsiella pneumoniae successfully treated with perfloxacin in neonate. Infection 1994;22:60-2.
- 22. Haley R. W., Cushion N. B., Tenover F. C., et al. Eradication of endemic methicillin-resistant Staphylococcus aureus infections from a neonatal intensive care unit. J Infect Dis 1995;171:614-24.
- 23. Loureiro M.M., de Moraes B.A., Quadra M.R.R., et al. Molecular Epidemiology of Methicillin Resistant Staphylococcus aureus Isolated from Newborns in a Hospital in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz, Rio de Janeiro 2000;95:777-82.
Publication Dates
-
Publication in this collection
21 Nov 2003 -
Date of issue
Apr 2003
History
-
Reviewed
13 May 2002 -
Received
08 Mar 2002