Abstract
We performed a comparative analysis of the nucleolus organizer region (NOR) distribution in the karyotypes of hylid frogs from two different taxonomic groups, Hypsiboas faber and H. semilineatus. Silver nitrate staining of NORs (Ag-NORs) and fluorescence in situ hybridization (FISH) with a rDNA probe were used to investigate the chromosomal location of rDNA loci in two species. The karyotype of H. semilineatus and the Ag-NORs distribution of the four species are presented herein for the first time. After conventional staining, the four species presented very similar karyotypes with 2n = 24, but Ag-NORs analyses revealed species-specific characteristics. H. albomarginatus, H. faber and H. semilineatus had one pair of interstitial Ag-NORs in the short arm of pairs 2, 11, and in the long arm of pair 7, respectively. H. pardalis presented telomeric NORs in the long arm of pair 11. Ag-NORs were heteromorphic in three of the species ( H. pardalis , H. semilineatus and H. albomarginatus ) and FISH confirmed the differential activity of rDNA genes in H. semilineatus . In the present study, 2n = 24 karyotypes could be distinguished by their Ag-NORs distribution. Our results further the knowledge about the cytogenetics of hylids from Brazil.
hylids; Hypsiboas; karyotypes; FISH; Ag-NORs; Hypsiboas semilineatus; H. faber
GENETICS
Patterns of ribosomal DNA distribution in hylid frogs from the Hypsiboas faber and H. semilineatus species groups
Rosana dos Reis Abrante Nunes
Valéria Fagundes
1Departamento de Ciências Biológicas, Centro de Ciências Humanas e Naturais, Universidade Federal do Espírito Santo, Vitória, ES, Brazil
We performed a comparative analysis of the nucleolus organizer region (NOR) distribution in the karyotypes of hylid frogs from two different taxonomic groups, Hypsiboas faber and H. semilineatus. Silver nitrate staining of NORs (Ag-NORs) and fluorescence in situ hybridization (FISH) with a rDNA probe were used to investigate the chromosomal location of rDNA loci in two species. The karyotype of H. semilineatus and the Ag-NORs distribution of the four species are presented herein for the first time. After conventional staining, the four species presented very similar karyotypes with 2n = 24, but Ag-NORs analyses revealed species-specific characteristics. H. albomarginatus, H. faber and H. semilineatus had one pair of interstitial Ag-NORs in the short arm of pairs 2, 11, and in the long arm of pair 7, respectively. H. pardalis presented telomeric NORs in the long arm of pair 11. Ag-NORs were heteromorphic in three of the species (H.pardalis, H. semilineatus and H. albomarginatus) and FISH confirmed the differential activity of rDNA genes in H. semilineatus. In the present study, 2n = 24 karyotypes could be distinguished by their Ag-NORs distribution. Our results further the knowledge about the cytogenetics of hylids from Brazil.
Anuran amphibians are frequently considered as a karyologically conservative group. In the family Hylidae, a basic karyotype with 2n = 24, morphologically similar chromosomes and no differentiated sex chromosomes has been described. About 25% of the known anuran species had their karyotype described until the 90s (Kuramoto, 1990; King, 1990), but the studies were mainly based on conventional staining analyses providing insufficient data to characterize species of hylids. More recent studies, which included chromosome banding (mainly C-banding and BrdU incorporation) and localization of the nucleolus organizer regions (NORs), revealed some minor differences among species, which became important tools in comparative chromosome analyses (Baldissera et al., 1993; Schmid et al., 1995; Silva et al., 2000; Kasahara et al., 2003; Ananias et al., 2004; Raber et al., 2004).
Interindividual variation and size heteromorphisms of the NORs detected by silver nitrate staining (Ag-NORs) frequently allow the karyological distinction of hylids. According to Schmid (1978Schmid M. (1978) Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 66:361-388.; 1982Schmid M. (1982) Chromosome banding in Amphibia VII: Analysis of the structure and variability of NORs in Anura. Chromosoma 87:327-344.), the Ag-NORs frequently occur at the same chromosomal location in the karyotypes of species of the same group or in groups of related species, reinforcing the importance of extensive karyological analyses to characterize the karyotypes of the group.
In this work, we aimed to compare the karyotypes and Ag-NORs patterns of four hylid species from the Atlantic Forest of southeastern Brazil. The species studied belong to two groups: the H. semilineatus group (H. semilineatus) and the H. faber group (H. albomarginatus, H. faber and H. pardalis). We also intended to verify if the Ag-NORs patterns were consistent with the monophyletic clades identified by molecular and morphological traits. We expected each species to present a distinctive Ag-NORs distribution which would be useful to distinguish individual species or species groups of hylids.
Our sample consisted of 13 specimens of species included in the hylid species groups H. semilineatus (H.semilineatus) and H. faber (H. albomarginatus, H. faber and H. pardalis). The animals were collected in four localities of the state of Espírito Santo, Brazil (Table 1). They were treated with a solution of 0.1% colchicine (1 mL/100 g of body weight) during 4-6 h prior to the sacrifice. Mitotic chromosome preparations were obtained from the intestine by the squash technique described by Bogart (1973a). Briefly, the intestine was immersed for 30 min in distilled water and then fixed in an ethanol/acid acetic solution. The intestine was fragmented with a scalpel onto a glass slide with a few drops of the fixative and then squashed with a glass coverslip. The slide was immersed in liquid nitrogen for about one minute and the coverslip was removed. The slide was then immersed in 90% ethanol for about one minute and air-dried. For conventional staining, the chromosome preparations were hydrolyzed in HCl 1N at 60 °C for 10 min and stained with a 3% Giemsa solution for 10 min. Ag-NORs staining followed Howell and Black (1980). Metaphases of each species were analyzed to determine the diploid number (2n) and the number of chromosome arms (fundamental number, FN). The metaphases were photographed and copies were made in Kodak photographic paper. Ten metaphases per species had their chromosomes measured (in millimeters) to determine the chromosome relative length (RL) and the centromeric ratio (CR), i.e. the proportion between the short and the long arms. We adopted the nomenclature for chromosome morphology proposed by Green and Sessions (1991), which is based on the centromeric ratio. Fluorescence in situ hybridization (FISH) was performed with the biotin-labeled probe HM123, which contains fragments of the 18S and 28S rDNA of Xenopus leavis (Meunier-Rotival et al., 1979). H. semilineatus and H. pardalis metaphases were hybridized with the probe following the protocol described by Viégas – Péquignot (1992).
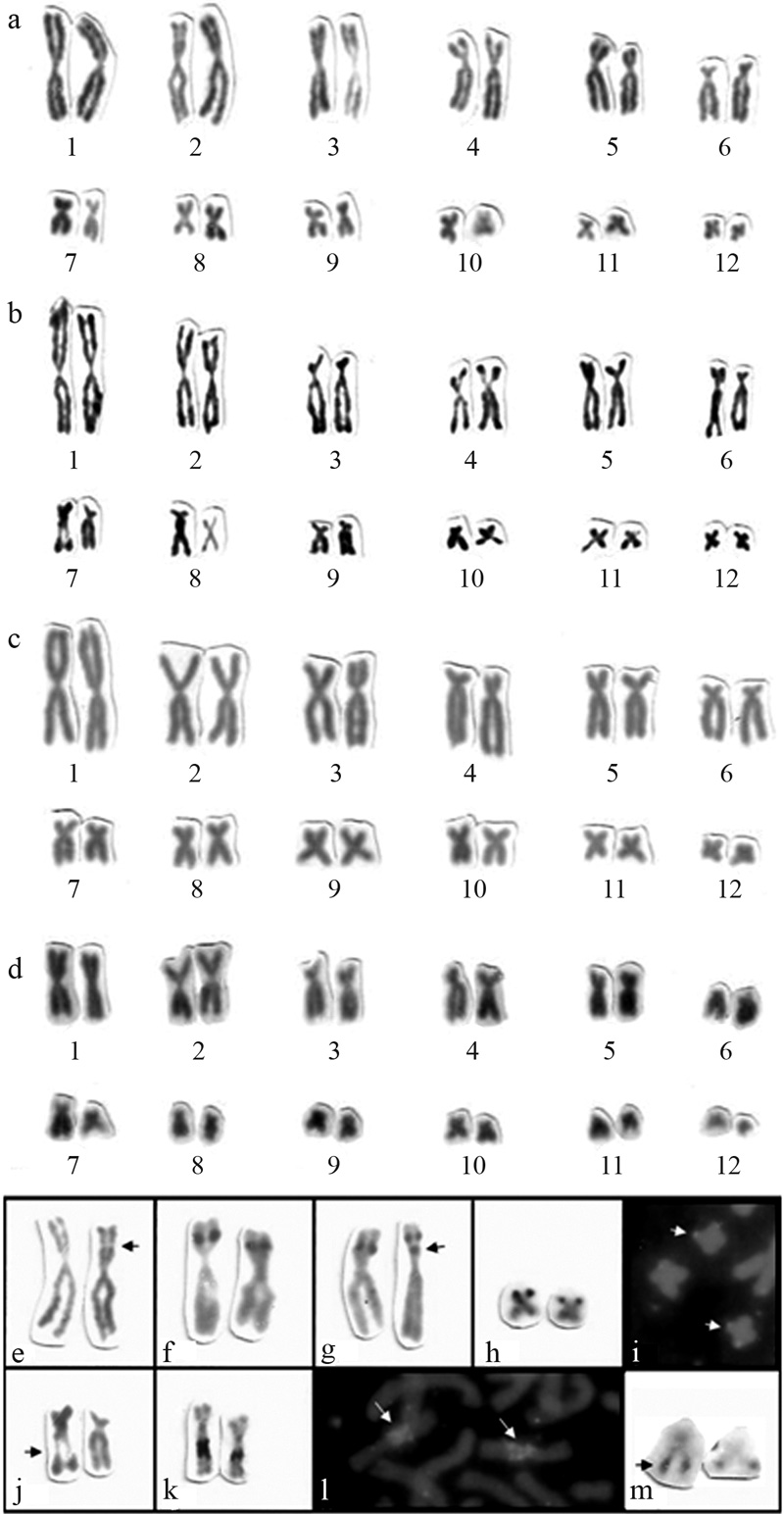
All specimens exhibited 2n = 24 and FN = 48 (Figure 1a-d), but comparative analyses revealed some differences in the relative length (RL) and chromosome morphologies among the karyotypes of the four species, even though they were all exclusively composed of biarmed chromosomes (Figure 1). No sex chromosomes were observed in any of the karyotypes. The chromosomes varied gradually in size, with pair 1 corresponding to 18-19% and pair 12 corresponding to 2-3% of the genome. Pair 6 of Hypsiboas faber was slightly smaller than the same chromosome pair in the other species (Table 2).
The karyotype of H. semilineatus (HSE) is described herein for the first time. It is composed of five pairs of metacentric chromosomes (pairs 1, 2, 10, 11 and 12), three pairs of submetacentrics (pairs 4, 5 and 8) and four pairs of subtelocentrics (pairs 3, 6, 7 and 9).
Hypsiboas albomarginatus (HAL) presented six pairs of metacentric chromosomes (pairs 1, 2, 3, 10, 11 and 12), five pairs of submetacentrics (pairs 4, 5, 7, 8 and 9) and one pair of subtelocentrics (pair 6). The karyotypes of H. albomarginatus from Espírito Santo were similar to those found in specimens from São Paulo identified as Hyla albomarginata by Beçak (1968).
H. pardalis (HPA) presented five pairs of metacentric chromosomes (pairs 1, 2, 9, 11 and 12), five pairs of submetacentrics (pairs 3, 5, 7, 8 and 10) and two pairs of subtelocentrics (pairs 4 and 6). The karyotype of Hyla pardalis from southeastern Brazil previously described by Bogart (1973b) is similar to the one that we observed of Hypsiboas pardalis from Espírito Santo.
H. faber (HFA) presented four pairs of metacentric chromosomes (pairs 1, 2, 9, and 12), four pairs of submetacentrics (pairs 3, 5, 8 and 10) and four pairs of subtelocentrics (pairs 4, 6, 7 and 11). The karyotype of Hyla faber from São Paulo described by Beçak (1968) is similar to that described herein for Hypsiboas faber.
A comparative analysis between the four species karyotypes revealed some conserved chromosome pairs (pairs 1, 2, 5, 6, 8, and 12), contrasting with some variable ones (pairs 3, 4, 7, 9, 10 and 11). Some more suggestive differences observed between homologues are more likely due to variations from the squashing technique than to real heteromorphisms. Pair 3 varied from subtelocentric (HSE) to submetacentric (HPA and HFA) and metacentric (HAL). Pair 4 was submetacentric (HSE and HAL) or subtelocentric (HPA and HFA). Pair 7 was submetacentric (HAL and HPA) or subtelocentric (HSE and HFA). Pair 9 was subtelocentric (HSE), submetacentric (HAL) or metacentric (HPA and HFA). Pair 10 was metacentric (HSE and HAL) or submetacentric (HPA and HFA) and pair 11 was metacentric (HSE, HAL and HPA) or subtelocentric (HFA).
Although a single chromosome pair presented an Ag-NOR in all metaphases from all the specimens, it was not the same pair in all species. Pairs 2 and 7 are the NORs-bearing chromosomes in H. albomarginatus and H. semilineatus, respectively, and present a secondary constriction, not always observed on both homologues (Figure 1e and 1j), which coincided with the location of rDNA gene clusters.
In H. albomarginatus, the Ag-NORs were interstitially located at the short arm of pair 2 (Figure 1e-f). In all metaphases both homologues had positive Ag-NORs signals. One specimen (LGA 221) exhibited an additional interstitial Ag-NOR, proximal to the short arm of one homologue of pair 2 (Figure 1g).
H. pardalis presented one pair of telomeric Ag-NORs in the short arms of pair 11 (Figure 1h). There was an exact correspondence in the position and size of the NORs after FISH and silver staining (Figure 1i).
All four specimens of H. semilineatus presented one interstitial Ag-NOR in the long arm of pair 7 (Figure 1j-k). A conspicuous difference in the size of the Ag-NORs between homologues was observed in all specimens. Despite of the heteromorphism in the Ag-NORs, the size of signals of the rDNA probes observed after FISH were similar on both homologues (Figure 1l).
Both specimens of H. faber presented interstitial Ag-NORs in the long arms of pair 11, which showed heteromorphism after silver-nitrate staining (Figure 1m).
Nucleolus organizer regions have been considered important markers for the study of chromosome evolution in amphibians (Lourenço et al., 1998). Although intraspecific variation of NORs location is rare (Schmid, 1978Schmid M. (1978) Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 66:361-388.), interindividual heteromorphisms of Ag-NORs size were widely observed in anurans. In an extensive study, Schmid, (1982)Schmid M. (1982) Chromosome banding in Amphibia VII: Analysis of the structure and variability of NORs in Anura. Chromosoma 87:327-344. used silver staining and GC-specific fluorochromes to analyze the NORs of 260 individuals from 23 genera of anurans. In this study, 67% of the animals presented heteromorphic Ag-NORs and in 69% of them the size differences could be attributed to tandem duplications or triplications affecting one of both rDNA clusters.
We observed Ag-NORs heteromorphisms in three of the four species investigated (H. semilineatus, H. albomarginatus and H. faber). The difference in the size of the Ag-NORs by silver staining of H. semilineatus was not confirmed by FISH with the rDNA probe, suggesting that the Ag-NORs heteromorphism is caused by differential gene activity (and consequent differential accumulation of ribonucleoproteins). The duplicated Ag-NORs sites in pair 2 of H. albomarginatus were also not tested by FISH. We were unable to determine if the Ag-NORs size heteromorphism observed in H. faber was due to variation in the number of ribosomal genes or to their differential activity, since FISH results were not obtained.
We observed duplicated Ag-NOR in H. albomarginatus. Duplicated Ag-NORs were previously reported by Schmid et al. (1995) in the frog Agalychnis callidryas. Possible mechanisms responsible for the dispersion of NOR sites in anuran genomes have been discussed by some authors (Wiley et al., 1989, King 1990, Foote et al., 1991, Schmid et al. 1995, Kaiser et al., 1996). They suggested that NORs transposition could have occurred by the movement of mobile genetic elements closely linked to NOR cistrons, amplifications of “orphan” rDNA cistrons, reinsertion errors during extrachromosomal amplification of ribosomal cistrons, or chromosomal rearrangements such as translocations and inversions involving the segments containing NORs. Oliveira et al. (1996) proposed that pericentric inversions could split NOR cistrons resulting in two Ag-NORs with about half of the regular size.
Considering the similar sizes of each of the duplicate Ag-NORs in H.albomarginatus, it is unlikely that pericentric inversions involving the rDNA region occurred in this species. We are thus inclined to attribute the duplication to an intrachromosomal duplication (likely due to mobile elements).
However, it is important to emphasize that the Ag-NOR technique detected heterochromatic regions in some species (Lourenço et al., 1998). Such regions were easily distinguished from the NORs sites by the intensity of silver impregnation, which resulted in black dots in true NORs and in brown sections in heterochromatic regions (Lourenço et al., 1998). We did not observe brownish staining in the extra signal, which allows us to discard that it is a silver-stained heterochromatic region. Moreover, the specimen with the heteromorphic duplicated Ag-NOR was collected in a locality different from that where the animals with non-duplicated Ag-NORs were found. New data on more individuals collected in the same locality would help to verify a possible regional variation.
Although the NORs location may be used to characterize species of amphibians, our analysis revealed that NORs are more useful to discriminate groups of species rather than species with similar 2n = 24 karyotypes. Anuran species frequently revealed only a single NOR-bearing chromosome pair in diploid karyotypes (Schmid, 1982Schmid M. (1982) Chromosome banding in Amphibia VII: Analysis of the structure and variability of NORs in Anura. Chromosoma 87:327-344.; Mahony and Robinson, 1986; Anderson, 1991). Schmid (1978Schmid M. (1978) Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 66:361-388.; 1982Schmid M. (1982) Chromosome banding in Amphibia VII: Analysis of the structure and variability of NORs in Anura. Chromosoma 87:327-344.) observed that the NORs usually occur at the same chromosomal location in the karyotypes of the species of the same group or in groups of related species. Exceptions to this rule indicate that rearrangements may have contributed to the species evolution (Schmid, 1978Schmid M. (1978) Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 66:361-388.).
We plotted our data on Ag-NORs location on Faivovich et al. (2005) phylogenetic tree and we observed that each monophyletic clade had the ribosomal cistron located in a specific chromosome pair (Figure 2).
H. pardalis (present study), H. faber (present study) and H. crepitans (Gruber et al., 2006), which belong to a monophyletic clade and are included in the H. faber group, shared the Ag-NORs at the pair 11. Differences of the relative size of chromosome 11, as specified for H. pardalis and H. faber in Table 2, are not rare. The variation in the NORs location (terminal in the short arm of a metacentric pair in H. pardalis, interstitial in the long arm of a subtelocentric pair in H. faber and interstitial in the long arm of a submetacentric pair in H. crepitans) can be explained by a single pericentric inversion in these chromosomes, changing the rDNA cluster from a telomeric to an interstitial position.
H. albomarginatus (present paper), also included in H. faber group, presented NORs in a different chromosome pair (pair 2). Although we only analyzed conventionally stained chromosomes we do not think that it is possible to misidentify pairs 2 and 11, and we believe that distinct chromosome pairs are NOR-bearing in each of these two clades.
In the H. pulchellus group, a sister group of H. faber, the species H. semiguttatus and H. joaquini (Ananias et al., 2004) grouped in a monophyletic clade and shared the presence of a telomeric Ag-NOR in pair 1. H. prasinus and H. pulchellus (Ananias et al., 2004) form another clade in the H. pulchellus group and shared the presence of Ag-NORs in pair 12. H. guentheri, H. bischoffi (Raber et al., 2004) and H. marginatus (Ananias et al., 2004) are arranged in a third monophyletic clade species group and present Ag-NORs in pair 10, at the telomeric position of the long arm of H. guentheri and H. bischoffi, and at the pericentromeric region in H. marginatus. These differences in NORs location can be explained by pericentric inversion.
Schmid (1978Schmid M. (1978) Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 66:361-388.; 1982Schmid M. (1982) Chromosome banding in Amphibia VII: Analysis of the structure and variability of NORs in Anura. Chromosoma 87:327-344.) stated that the NORs were recurrently observed at the same chromosomal location in the karyotypes of species from the same groups and that chromosome translocations could be frequent in the evolution of the group. Our data corroborate this assumption and suggest that: (1) inversions also contributed to chromosome changes and NORs transpositions; (2) Ag-NORs present a pattern in hylid frogs species groups (as observed in the H. faber group) or inside clades of species groups (as in the H. pulchellus group). These data may be useful in the cytotaxonomic and evolutionary studies of hylids.
a-d) Karyotypes with 2n = 24, FN = 48 after conventional staining: a) Hypsiboas albomarginatus, b) H. semilineatus, c) H. pardalis and d) H. faber; e-g) Pair 2 in H. albomarginatus: e) secondary constriction (arrow), f) homomorphic Ag-NORs at the short arms, g) heteromorphic Ag-NORs with an extra NOR in one homologue (arrow); h-i) Pair 11 of H. pardalis: h) homomorphic Ag-NORs, terminal in the short arms, i) homomorphic FISH signals with rDNA probes (arrows); j-l) Pair 7 in H. semilineatus: j) secondary constriction (arrow), k) heteromorphic Ag-NORs, l) homomorphic FISH signals with rDNA probes (arrows); m) Pair 11 of H. faber with interstitial heteromorphic Ag-NORs in the long arms (arrow).
Figure 2 Partial view of the maximum parsimony consensus tree presented by Faivovich et al. (2005), modified with the inclusion of our Ag-NORs data. Wherever NOR data are missing, this is due to the absence of data in the literature.
Acknowledgments
This work was supported by the Conselho Nacional Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, RRAN Masters scholarship). We thank the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis for the license to collect (IBAMA n. 02009.00-1492/00, license 021/2000 DIFAS), and Pedro Luis Peloso and João Luís Gasparini for collecting and identifying some specimens. We are also grateful to Dr. S. Recco-Pimentel for providing the conditions in the laboratory to carry out the FISH experiments.
References
Ananias F., Garcia P.C.A., Recco-Pimentel S.M. (2004) Conserved karyotype in the Hyla pulchella species group (Anura, Hylidae). Hereditas 140:42-48.
Anderson K. (1991) Chromosome evolution in Holarctic Hyla treefrogs. Amphibian Cytogenetics and EvolutionGreen D.M., Sessions S.K.San DiegoAcademic Press299-328.
Baldissera F.A., Oliveira P.S.L., Kasahara S. (1993) Cytogenetics of four Brazilian Hyla species (Amphibia, Anura) and description of a case with a supernumerary chromosome. Rev Bras Genet 16:335-345.
Beçak M.L. (1968) Chromosomal analysis of eighteen species of Anura. Caryologia 21:191-208.
Bogart J.P. (1973a) Method for obtaining chromosomes. Caldasia 11:29-40.
Bogart J.P. (1973b) Evolution of anuran karyotypes. Evolutionary Biology of the AnuransVial J.L.ColumbiaUniversity of Missouri Press337-349.
Faivovich J., Haddad C.F.B., Garcia P.C.A., Frost D.R., Campbell J.A., Wheeler W.C. (2005) Systematic review of the frog family Hylidae, with special reference to Hylinae: Phylogenetic analysis and taxonomic revision. Bull Am Mus Nat Hist 294:1-240.
Foote D.L., Wiley J.E., Little M.L., Meyne J. (1991) Ribosomal RNA gene site polymorphism in Bufo terrestris. Cytogenet Cell Genet 57:196-199.
Green D.M., Sessions S.K. (1991) Nomenclature for chromosomes. Amphibian Cytogenetics and EvolutionGreen D.M., Sessions S.K.San DiegoAcademic Press431-432. Gruber S.L., Haddad C.F.B., Kasahara S. (2006) Chromosome banding in three species of Hypsiboas (Hylidae, Hylinae), with special reference to a new case of B-chromosome in anuran frogs and to the reduction of the diploid numbers of 2n = 24 to 2n = 22 in the genus. Genetica 130:281-229.Howell W.M., Black D.A. (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1- step method. Experientia 36:1014-1015.
Kaiser H., Mais C., Bolanos F., Steinlein C., Feichtinger W., Schmid M. (1996) Chromosomal investigation of three Costa Rica frogs from the 30-chromosome radiation of Hyla with the description of a unique geographic variation in nucleolus organizer regions. Genetica 98:95-102.
Kasahara S., Silva A.P.Z., Gruber S.L., Haddad C.F.B. (2003) Comparative cytogenetic analysis on four tree frog species (Anura, Hylidae, Hylinae) from Brazil. Cytogenet Gen Res 103:155-162.
King M. (1990) Amphibian. Animal CytogeneticsJohn B.Berlin and StuttgartGebruder Borntraegger1-241.
Kuramoto M. (1990) A list of chromosome number of anuran amphibians. Bull Fukoka Univ Educ 39:83-127.
Lourenço L.B., Recco-Pimentel S.M., Cardoso A.J. (1998) Polymorphism of the nucleolus organizer regions (NORs) in Physalemus petersi (Amphibia, Anura, Leptodactylidae) detected by silver staining and fluorescence in situ hybridization. Chromosome Res 6:621-628.
Mahony M.J., Robinson E.S. (1986) Nucleolar organiser region (NOR) location in karyotypes of Australian ground frogs (family Myobatrachidae). Genetica 68:119-127.
Meunier-Rotival M., Cortadas J., Macaya G., Bernardi G. (1979) Isolation and organization of calf ribosomal DNA. Nucleic Acids Res 6:2109-2123.
Oliveira C., Foresti F., Tabata Y.A. (1996) Pericentric inversion involving a NOR bearing chromosome of rainbow trout (Oncorhynchus mykiss): Electron microscopy studies of the synaptonemal complex. Caryologia 49:335-342.
Raber S.C., Carvalho K.A., Garcia P.C.A., Vinciprova G., Recco-Pimentel S.M. (2004) Chromosomal caracterization of Hyla bischoffi and Hyla guentheri (Anura, Hylidae). Phylomedusa 3:43-49.
Schmid M., Feichtinger W., Weimer R. (1995) Chromosome banding in Amphibia XXI. Inversion polymorphism and nucleolus organizer regions in Agalychnis callidryas (Anura, Hylidae). Cytogenet Cell Genet 69:18-26.
Silva A.P.Z., Baldissera Jr F.A., Haddad C.F.B., Kasahara S. (2000) Karyotypes and nucleolus organizer regions of the genus Physalaemus (Anura, Leptodactylidae). Iheringia Ser Zool 88:159-164.
Viegas-Péquignot E. (1992) In situ hybridization to chromosomes with biotinylated probes. In situ Hybridization: A Practical ApproachWillerson D.OxfordOxford University Press and IRL Press137-158.
Wiley J.E., Little M.L., Romano M.A., Blount D.A., Cline G.R. (1989) Polymorphism in the location of the 18S and 28S rRNA genes on the chromosomes of the diploid-tetraploid tree frogs Hylachrysoscelis and Hylaversicolor. Chromosoma 97:481-487.
1M = Male; ND = not determined; 2CS – number of cells analyzed after conventional staining; Ag-NORs – number of cells analyzed after Ag-NOR staining.
- Relative chromosome length (RL), centromeric ratio (CR) and morphology (CP) of the mitotic chromosomes of Hypsiboas albomarginatus (HAL), H. semilineatus (HSE), H. pardalis (HPA) and H. faber (HFA).
m = metacentric; sm = submetacentric and st = subtelocentric.
Received: February 18, 2008; Accepted: June 17, 2008
Valéria Fagundes. Laboratório de Genética Animal, Departamento de Ciências Biológicas, Centro de Ciências Humanas e Naturais, Universidade Federal do Espírito Santo, Av. Marechal Campos 1468, Maruípe, 29040-90 Vitória, ES, Brazil. E-mail: vfagunde@npd.ufes.br or vfagunde@pesquisador.cnpq.br.
Publication Dates
-
Publication in this collection
19 Nov 2008 -
Date of issue
2008
History
-
Received
1802 -
Accepted
1706