Abstract
Urease catalyzes the hydrolysis of urea to ammonia and carbon dioxide. The ammonia (nitrogen (N) product of urease activity) is incorporated into organic compounds. Thus, urease is involved in N remobilization, as well as in primary N assimilation. Two urease isoforms have been described for soybean: the embryo-specific, encoded by the Eu1 gene, and the ubiquitous urease, encoded by Eu4. A third urease-encoding gene was recently identified, designated Eu5, which encodes the putative protein product SBU-III. The present study aimed to evaluate the contribution of soybean ureases to seed germination and plant development. Analyses were performed using Eu1/Eu4/Eu5-co-suppressed transgenic plants and mutants of the Eu1 and Eu4 urease structural genes, as well as a urease-null mutant (eu3-a) that activates neither the ubiquitous nor embryo-specific ureases. The co-suppressed plants presented a developmental delay during the first month after germination; shoots and roots were significantly smaller and lighter. Slower development was observed for the double eu1-a/eu4-a mutant and the eu3-a single mutant. The N content in transgenic plants was significantly lower than in non-transgenic plants. Among the mutants, eu3-a presented the lowest and eu1-a the highest N content. Altogether, these results indicate that increased ureolytic activity plays an important role in plant development.
Keywords:
Plant development; functional study; transgenic plants; mutants; urease
Introduction
Nitrogen (N) is the most limiting plant nutrient, possibly after fixed carbon, for plant growth and development (Marschner, 2012Marschner P (2012) Marschner's Mineral Nutrition of Higher Plants. 3rd edition. Academic Press, London, 651p.). Therefore, efficient mechanisms both to take up N in its various forms and to reallocate it are necessary for optimal N use efficiency (Witte, 2011Witte CP (2011) Urea metabolism in plants. Plant Sci 180:431-438.). In plant cells, urea is an important internal and external source of N that must be converted to ammonia for N assimilation (Wang et al., 2008Wang WH, Kohler B, Cao FQ and Liu LH (2008) Molecular and physiological aspects of urea transport in higher plants. Plant Sci 175:467-477.). In nature, two major biochemical processes lead to urea production: (1) arginase-catalyzed production of urea (and ornithine) from arginine (a major N storage form); (2) purine degradation to glyoxylate and urea. While arginases are active in plants, purine degradation exclusively through urea, though occurring in many bacteria, fungi and algae, does not occur in soybean and Arabidopsis. Rather, in these dicotyledonous plants, the purine degradation product allantoin, which contains the four ring N atoms of purine, is degraded to four ammonia molecules, bypassing a urea intermediate (summarized in Witte, 2011Witte CP (2011) Urea metabolism in plants. Plant Sci 180:431-438.).
Nonetheless, plant assimilation of arginine-derived urea is important for efficient N use, both in mobilization of seed N reserves during germination, as well as in remobilization of N in senescing tissues. In soybean, arginine is the major amino acid repository of seed N (Micallef and Shelp, 1989Micallef BJ and Shelp BJ (1989) Arginine metabolism in developing soybean cotyledons. I. Relationship to nitrogen nutrition. Plant Physiol 90:624-630.), and one of the predominant amino acids in angiosperm seed protein in general (Van Etten et al., 1963Van Etten CH, Miller RW, Wulf IA and Jones Q (1963) Amino acid composition of seeds from 200 angiospermous plant species. J Agric Food Chem 11:399-410.). In soybean, arginase action during germination releases much urea, which is hydrolyzed by urease action (Goldraij and Polacco, 1999Goldraij A and Polacco JC (1999) Arginase is inoperative in developing soybean seeds. Plant Physiol 119:297-304., 2000Goldraij A and Polacco JC (2000) Arginine degradation by arginase in mitochondria of soybean seedling cotyledons. Planta 210:652-658.)
The importance of urease for recycling N was highlighted in aged Arabidopsis thaliana seeds that failed to germinate when urease was chemically inhibited, but could be rescued by an external N source (Zonia et al., 1995Zonia LE, Stebbins N and Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107:1097-1103.). And, according to Bohner et al. (2015)Bohner A, Kojima S, Hajirezaei M, Melzer M and Von Wiren N (2015) Urea retranslocation from senescing Arabidopsis leaves is promoted by DUR3-mediated urea retrieval from leaf apoplast. Plant J 81:377-387. 13% of N exported out of senescing leaves of A. thaliana via the petiole is urea. Urease-negative soybean (mutants and nickel-deprived wildtype) accumulate urea in necrotic leaf tips (Stebbins et al., 1991Stebbins N, Holland MA, Cianzio SR and Polacco JC (1991) Genetic tests of the roles of the embryonic ureases of soybean. Plant Physiol 97:1004-1010.) to levels approaching 2.5% dry weight (Eskew et al., 1983Eskew DL, Welch RM and Cary EE (1983) Nickel: An essential micronutrient for legumes and possibly all higher plants. Science 222:621-623.). We note that in Stebbins, (1991)Stebbins N, Holland MA, Cianzio SR and Polacco JC (1991) Genetic tests of the roles of the embryonic ureases of soybean. Plant Physiol 97:1004-1010. and Eskew et al. (1983)Eskew DL, Welch RM and Cary EE (1983) Nickel: An essential micronutrient for legumes and possibly all higher plants. Science 222:621-623. available N was not limiting.
Urea can be hydrolyzed by two different enzymes: urease and an ATP (and biotin)-dependent urea carboxylase/allophanate hydrolase. The latter, found in some fungi, algae and at least one bacterium (Kanamori et al., 2004Kanamori T, Kanou N, Atomi H and Imanaka T (2004) Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol 186:2532-2539.), has never been reported in plants. Rather, all plants appear to have a urease (Hogan et al., 1983Hogan ME, Swift IE and Done J (1983) Urease assay and ammonia release from leaf tissues. Phytochemistry 22:663-700. and our own observations). In soybean and Arabidopsis, urea nitrogen is only available after urea hydrolysis by urease (Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.; Witte, 2011Witte CP (2011) Urea metabolism in plants. Plant Sci 180:431-438.; Polacco et al., 2013Polacco JC, Mazzafera P and Tezotto T (2013) Opinion - Nickel and urease in plants: Still many knowledge gaps. Plant Science 199-200:79-90.). Urease (EC 3.5.1.5) was the first identified nickel-dependent metalloenzyme (Dixon et al., 1975Dixon NE, Gazzola TC, Blakeley RL and Zermer B (1975) Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J Am Chem 97:4131-4133.), and much has been learned of the construction and function of its metallocenter (Carter et al., 2009Carter EL, Flugga N, Boer JL, Mulrooney SB and Hausinger RP (2009) Interplay of metal ions and urease. Metallomics 1:207-221.). Urease catalyzes the hydrolysis of urea to ammonia and carbon dioxide. In addition to plants, ureases are synthesized by bacteria, fungi and algae (Krajewska, 2009Krajewska B (2009) Ureases I. Functional, catalytic and kinetic properties: A review. J Mol Catal B Enzymatic 59:9-21.). The N product of urease activity - ammonia - is incorporated into organic compounds mainly by glutamine synthase activity (Mobley et al., 1995Mobley HL, Island MD and Hausinger RP (1995) Molecular biology of microbial ureases. Microbiol Rev 59:451-480.; Sirko and Brodzik, 2000Sirko A and Brodzik R (2000) Plant ureases: Roles and regulation. Acta Biochim Pol 47:1189-1195.). Thus, urease is involved in N remobilization, as well as in primary N assimilation (Cao et al., 2010Cao F-Q, Werner AK, Dahncke K, Romeis T, Liu L-H and Witte CP (2010) Identification and characterization of proteins involved in rice urea and arginine catabolism. Plant Physiol 15:98-108.).
In addition to the N assimilatory function of urease, plant ureases appear to have defensive roles against herbivore and fungal attack (Carlini and Ligabue-Braun, 2016Carlini C and Ligabue-Braun R (2016) Ureases as multifunctional toxic proteins: A review. Toxicon 110:90-109.). In soybean, three urease isoforms have been described. The ubiquitous urease, encoded by the Eu4 gene, is expressed at low levels in all tissues and is responsible for recycling both metabolically-derived and exogenous urea (Polacco et al., 1985Polacco JC, Krueger RW and Winkler RG (1985) Structure and possible ureide degrading function of the ubiquitous urease of soybean. Plant Physiol 79:794-800.; Torisky et al., 1994Torisky RS, Griffin JD, Yenofsky RL and Polacco JC (1994) A single gene (Eu4) encodes the tissue-ubiquitous urease of soybean. Mol Gen Genet 242:404-414.; Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.). The embryo-specific urease, encoded by Eu1, is highly expressed in developing embryos and accumulates in mature seeds (Polacco and Havir, 1979Polacco JC and Havir EA (1979) Comparisons of soybean urease isolated from seed and tissue culture. J Biol Chem 254:1707-1715.; Polacco and Winkler, 1984Polacco JC and Winkler RG (1984) Soybean leaf urease: A seed enzyme? Plant Physiol 74:800-803.; Polacco and Holland, 1993Polacco JC and Holland MA (1993) Roles of urease in plant cells. Int Rev Cytol 47:1189-1195.). A third urease-encoding gene was recently identified in the soybean genome (Polacco et al., 2011Polacco JC, Hyten DL, Medeiros-Silva M, Sleper DA and Bilyeu KD (2011) Mutational analysis of the major soybean UreF paralogue involved in urease activation. J Exp Bot 62:3599-3608., 2013Polacco JC, Mazzafera P and Tezotto T (2013) Opinion - Nickel and urease in plants: Still many knowledge gaps. Plant Science 199-200:79-90.). This gene was designated Eu5, and its putative protein product was named SBU-III. Eu5 is expressed in the first stages of root development and during seed maturation. Its transcript levels are lower than those of the other two soybean urease isoforms (Wiebke-Strohm et al., 2016Wiebke-Strohm B, Ligabue-Braun R, Rechenmacher C, de Oliveira-Busatto LA, Carlini CR and Bodanese-Zanettini MH (2016) Structural and transcriptional characterization of a novel member of the soybean urease gene family. Plant Physiol Biochem 101:96-104.).
Urease-negative mutant soybean plants were examined to ascertain the role(s) of the urease isoforms. An embryo-specific urease null mutant (eu1-a) seems not to exhibit an altered physiology (Polacco et al., 2011Polacco JC, Hyten DL, Medeiros-Silva M, Sleper DA and Bilyeu KD (2011) Mutational analysis of the major soybean UreF paralogue involved in urease activation. J Exp Bot 62:3599-3608.). Ubiquitous urease missense mutants (eu4-a and eu4-b) produce an inactive protein and display no ureolytic activity in leaves, roots and hypocotyls (Torisky and Polacco, 1990Torisky RS and Polacco JC (1990) Soybean roots retain the seed urease isozyme synthesized during embryo development. Plant Physiol 94:681-689.; Stebbins et al., 1991Stebbins N, Holland MA, Cianzio SR and Polacco JC (1991) Genetic tests of the roles of the embryonic ureases of soybean. Plant Physiol 97:1004-1010.; Witte et al., 2002Witte CP, Tiller SA, Taylor MA and Davies HV (2002) Leaf urea metabolism in potato. Urease activity profile and patterns of recovery and distribution of (15) N after foliar urea application in wild-type and urease-antisense transgenics. Plant Physiol 128:1129-1136., Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.; Polacco et al., 2011Polacco JC, Hyten DL, Medeiros-Silva M, Sleper DA and Bilyeu KD (2011) Mutational analysis of the major soybean UreF paralogue involved in urease activation. J Exp Bot 62:3599-3608.). eu4 callus cultures cannot use 5 mM urea as N source, but are resistant to 50 mM urea in the presence of a standard NH4+ + NO3- N source, and show growth responses contrary to those of Eu4 cultures (Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.). The eu1-a/eu4-a double mutants were considered virtually devoid of ureolytic activity (Stebbins and Polacco, 1995Stebbins N and Polacco JC (1995) Urease is not essential for ureide degradation in soybean. Plant Physiol 109:169-175.; Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.).
A null mutant for the Eu3 gene, which encodes an accessory protein, UreG, necessary for urease activation, has also been characterized (Freyermuth et al., 2000Freyermuth SK, Bacanamwo M and Polacco JC (2000) The soybean Eu3-a gene encodes an Ni-binding protein necessary for urease activity. Plant J 21:53-60.). There is only a single copy of this gene in the soybean genome, and the deletion mutant eu3-a exhibits a complete loss of urease activity (Stebbins and Polacco, 1995Stebbins N and Polacco JC (1995) Urease is not essential for ureide degradation in soybean. Plant Physiol 109:169-175.; Polacco et al., 2011Polacco JC, Hyten DL, Medeiros-Silva M, Sleper DA and Bilyeu KD (2011) Mutational analysis of the major soybean UreF paralogue involved in urease activation. J Exp Bot 62:3599-3608.; Tezotto et al., 2016Tezotto T, Souza SCR, Mihail J, Favarin JL, Mazzafera P, Bilyeu K and Polacco JC (2016) Deletion of the single UreG urease activation gene in soybean NIL lines: Characterization and pleiotropic effects. Theor Exp Plant Physiol 28:307-320.).
A previous study was performed by our team aiming to overexpress Eu4 in soybean plants. Unexpectedly, the transgenic plants exhibited co-suppression of the endogenous and the introduced Eu4 transgene, resulting in decreased ureolytic activity (Wiebke-Strohm et al., 2012Wiebke-Strohm B, Pasquali G, Margis-Pinheiro M, Bencke M, Bücker-Neto L, Becker-Ritt AB, Martinelli AHS, Rechenmacher C, Polacco JC, Stolf R, et al. (2012) Ubiquitous urease affects soybean susceptibility to fungi. Plant Mol Biol 78:1-13.). As null mutants for the ubiquitous urease have not been obtained to date, the co-suppressed transgenic plants represent a powerful tool for functional gene studies. Here, we sought to determine the roles of urease in soybean development by elimination of all urease isoforms.
Material and Methods
Plant material and growth conditions
Homozygous eu1-a, eu4-a, eu1-a/eu4-a and eu3-a mutants have been described previously. All, except eu1-a, were recovered from EMS (ethyl methane sulfonate) mutagenesis of cv. Williams, and were subsequently outcrossed to Wiliams 82. The original eu1-a mutation was recovered from the “Itachi' landrace and introgressed into Williams by Dr Dick Bernard (University of Illinois-Champaign-Urbana) by five crosses. Thus, the genetic background of these mutants is the Williams (eu1-a) and Williams82 (eu4-a, eu1-a/eu4-a and eu3-a) cultivars. Williams and Williams82 are supposedly isogenic, except for a fungal resistance gene introgressed into Williams 82 (Bernard and Cremeens, 1988Bernard RL and Cremeens CR (1988) Registration of Williams 82 soybean. Crop Sci 28:1027-1028.).
Two independent transgenic events (A3 and A8) of soybean cultivar IAS5 that presented co-suppression of eu4 were obtained from bombarded embryogenic tissue. The vector used for transformation contained the Eu4 and the gfp-encoding sequences (Wiebke-Strohm et al., 2012Wiebke-Strohm B, Pasquali G, Margis-Pinheiro M, Bencke M, Bücker-Neto L, Becker-Ritt AB, Martinelli AHS, Rechenmacher C, Polacco JC, Stolf R, et al. (2012) Ubiquitous urease affects soybean susceptibility to fungi. Plant Mol Biol 78:1-13.). Plants derived from non-transgenic embryogenic tissues submitted to the same culture conditions were recovered and used as a control. Subsequent generations were obtained by self-fertilization of plants.
Transgenic seeds of A3 and A8 events (from T1, T2 and T3) were placed in Petri dishes containing sterile filter paper moistened with sterile distilled water for 24 h. Seeds expressing the gfp reporter were selected under blue light using a fluorescence stereomicroscope (Olympus®), equipped with a BP filter set with a 488 nm excitation filter and a 505-530 nm emission filter. GFP-positive and negative plants were also PCR-screened to confirm presence/absence of the transgene using the protocol described by Wiebke-Strohm et al. (2012)Wiebke-Strohm B, Pasquali G, Margis-Pinheiro M, Bencke M, Bücker-Neto L, Becker-Ritt AB, Martinelli AHS, Rechenmacher C, Polacco JC, Stolf R, et al. (2012) Ubiquitous urease affects soybean susceptibility to fungi. Plant Mol Biol 78:1-13.. Positive transgenic, as well as non-transgenic seeds, were sown in organic soil and maintained in a greenhouse until maturity at FUNDACEP-CCGL (Cruz Alta, RS, Brazil) and supplemented with a nutrient solution containing either NO3 or NH4 (as N source).
For seed germination and developmental evaluation, GFP-positive transgenic (T2), mutants, IAS5 non-transgenic and Williams82 seeds were sown in pots containing vermiculite and maintained for one month in a growth chamber at 26 ± 1° C with a 16/8 h light/dark cycle at a light intensity of 22.5 μEm-2s-1. Plants were not supplemented with any nutrient solution during the first 30 days of development.
RNA extraction, cDNA synthesis and quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from roots with Trizol reagent (Invitrogen) and treated with DNase I (Invitrogen) according to the manufacturers' instructions. First-strand cDNAs were obtained using 1 μg of DNA-free RNA, M-MLV Reverse Transcriptase SystemTM (Invitrogen) and oligo(dT) primers.
RT-qPCR was performed on a StepOne Real-time CyclerTM (Applied Biosystems). PCR-cycling conditions were implemented as described: 5 min at 94 °C, followed by 40 cycles of 10 s at 94 °C, 15 s at 60 °C and 15 s at 72 °C. A melting curve analysis was performed at the end of the PCR run, over the range of 55-99 °C, increasing the temperature stepwise by 0.1 °C every 1 s. Each 25-μL reaction comprised 12.5 μL cDNA (1:50 dilution), 1x PCR buffer (Invitrogen), 2.4 mM MgCl2, 0.024 mM dNTPs, 0.1 μM of each primer, 2.5 μL of SYBR-Green (1:100,000, Molecular Probes) and 0.03 U of Platinum Taq DNA Polymerase (5 U/μl, Invitrogen). All PCR assays were performed in technical quadruplicates and 10 biological samples. Reactions lacking cDNA were used as negative controls.
Transcript levels of the three urease-encoding genes were evaluated. The F-box protein and a metalloprotease were used as references for gene expression normalization (Jian et al,. 2008Jian B, Liu B, Bi Y, Hou W, Wu C and Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59.; Libault et al., 2008Libault MS, Thibivilliers DD, Bilgin O, Radwan M, Benitez SJ, Clough and Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1:44-54.). Primer sequences are presented in Table 1. The expression data analyses were performed after comparative quantification of amplified products using the 2-ΔΔCt method (Livak and Schmittgen, 2001Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408.).
Ureolytic activity
Urease activity in transgenic (A3 and A8) and non-transgenic plants was determined with a urease indicator solution: 6 g urea, 10 mL cresol red (1 mg/mL ethanol), 10 mL KH2PO4/K2HPO4/EDTA (10 mM KPi/1 mM EDTA pH 7.0) and 1 mL sodium azide 20% (w/v) per liter (Meyer-Bothling and Polacco, 1987Meyer-Bothling LE and Polacco JC (1987) Mutational analysis of the embryo-specific urease locus of soybean. Mol Gen Genet 209:439-444.). Powdered leaves and roots ( ± 100 mg) of two-week old plants, were incubated in a 1 mL urease indicator solution for 24 h at 60 °C, as well as mature seed slices that were incubated for 20 min at room temperature. The eu3-a mutant was used as negative control. As urea is hydrolyzed by urease, the ammonia released increases the pH and turns to pink the initial yellow coloration of the cresol red pH indicator.
Germination
Germination capacity of seeds (T2) from two transgenic events (A3 and A8) was compared with that of IAS5 non-transgenic seeds. For eu1-a, eu4-a, eu1-a/eu4-a and eu3-a mutant seeds germination capacity was compared with that of Williams82 non-mutant seeds. Seed germination, defined as radicle protrusion, was recorded over one month.
Developmental pattern
Seven, 14, 21 and 30 days after sowing, seedlings from all genotypes were classified according to their developmental stage following the categories proposed by Neumaier et al. (2000)Neumaier N, Nepomuceno AL and Farias JRB (2000) Estádios de desenvolvimento da cultura de soja. In: Bonato ER (ed). Estresses em Soja. EMBRAPA CNPT, Passo Fundo, pp 19-44.: VE = emergence of cotyledons; VC = completely opened cotyledons; V1 = completely developed unifoliate leaf pair; V2 = completely developed first trifoliate leaf; V3 = completely developed second trifoliate leaf. Eighteen plants derived from each event (A3 and A8), 10 non-transgenic plants, 18 plants from each mutant and 10 non-mutant plants were observed. In addition, the dry matter and length of roots and shoots were also determined one month after sowing.
Grain yield
The number of seeds produced per plant in three generations of transgenic (T1, T2 and T3) and non-transgenic plants were compared.
Nitrogen content
Shoot nitrogen content (N mobilized from the cotyledons) was measured by the Kjeldahl method according to the methodology described by Tedesco et al. (1995)Tedesco MJ, Gianello C, Bissani CA, Bohnen H and Vokweiss SJ (1995) Análise de Solo, Plantas e Outros Materiais. Boletim Técnico, 5. 3rd edition. UFRGS, Porto Alegre, 174 p. one month after sowing. This analysis evaluated 18 plants derived from each line A3 and A8 and 10 non-transgenic ones. Eighteen plants from each mutant (eu1-a, eu4-a, eu1-a/eu4-a and eu3-a) and non-mutant plants were also evaluated.
Statistical analysis
A Student's t-test was used to compare the expression levels (RT-qPCR) of urease-encoding genes (Eu4, Eu1, Eu5) in roots of transgenic vs. non-transgenic plants and mutant vs. non-mutant plants.
In order to compare developmental stages, a score was attributed to each developmental category (VE = 1, VC = 2, V1 = 3, V2 = 4, V3 = 5). A generalized linear model for repeated measures was used to compare plant development among genotypes (IAS5 transgenic vs. IAS5 non-transgenic plants; eu1-a or eu4-a or eu3-a or eu1-a/eu4-a mutant vs. Williams82 non-mutant plants). ANOVA followed by Bonferroni's post hoc test were performed on dry matter, length and weight of roots and shoots data. A Student's t-test was carried out in order to compare the number of seeds produced per transgenic and non-transgenic plant in different generations (T1, T2 and T3). Data on shoot nitrogen content was compared among genotypes by ANOVA followed by Tukey's post hoc test. Analyses were performed using SPSS 18.0 software.
Results
Gene expression
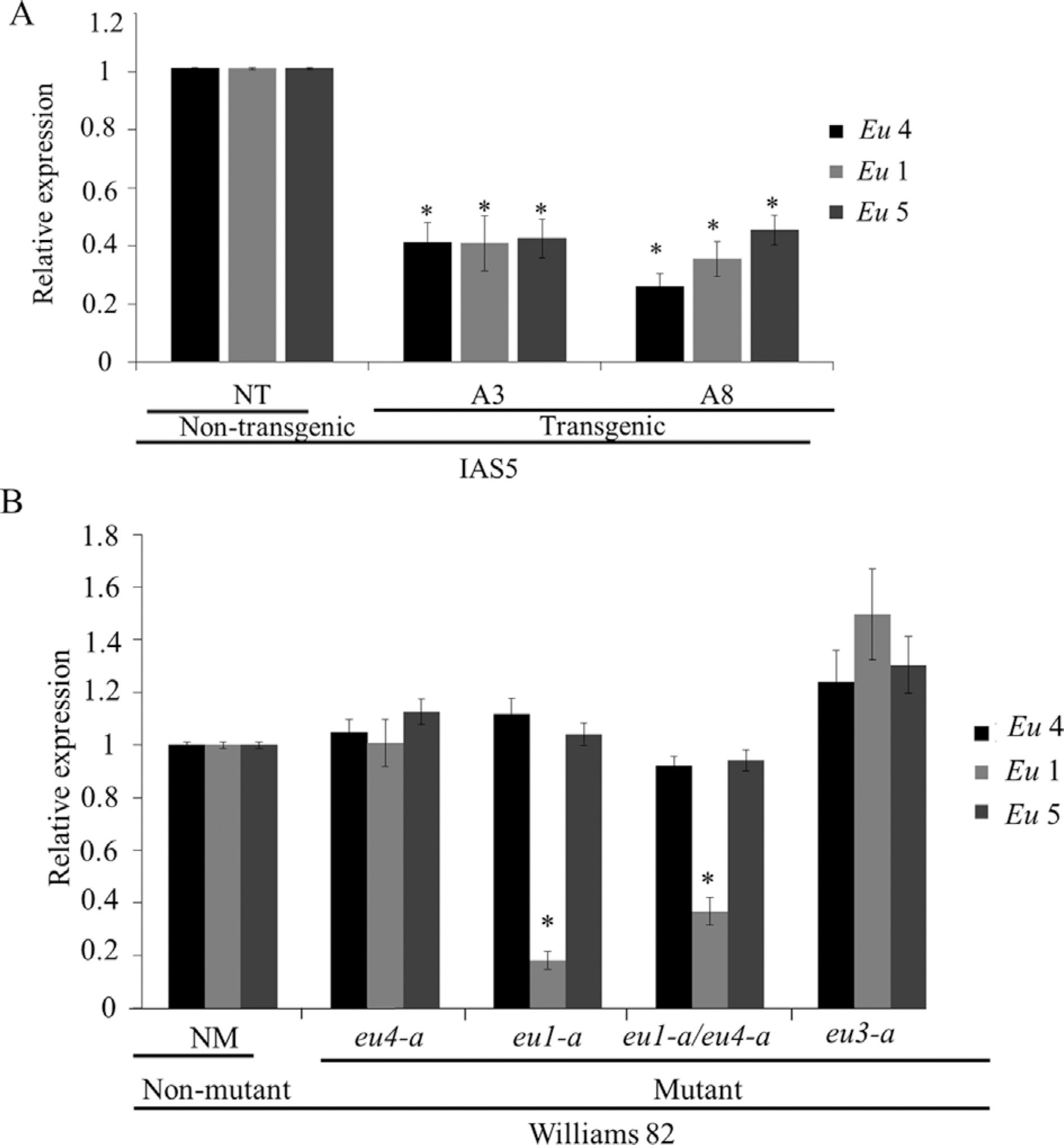
Roots were chosen for gene expression analysis by RT-qPCR because Eu5 transcripts are mainly detected in this organ (Wiebke-Strohm et al., 2016Wiebke-Strohm B, Ligabue-Braun R, Rechenmacher C, de Oliveira-Busatto LA, Carlini CR and Bodanese-Zanettini MH (2016) Structural and transcriptional characterization of a novel member of the soybean urease gene family. Plant Physiol Biochem 101:96-104.). As expected, the progeny of transgenic plants showed lower Eu4 transcript levels than non-transgenic controls, suggesting that the co-suppressed phenotype was maintained. Additionally, it was observed that the other urease-encoding genes, Eu1 and Eu5, were also down-regulated (Figure 1A).
Transcript levels (RT-qPCR) of urease-encoding genes (Eu4, Eu1, Eu5) in roots of two-week-old plants. (A) Two independent transgenic events (A3 and A8) and non-transgenic plants (NT, control) from cv. IAS5. (B) Williams82 non-mutant (NM, control) and eu4-a, eu1-a, eu1-a/eu4-a and eu3-a mutants. The bars represent mean ± SD of two non-transgenic plants, 10 transgenic plants from each event, two non-mutant plants and 10 plants from each mutant. Transcripts level of Eu4, Eu1 and Eu5 detected in non-transgenic or non-mutant plants were used to normalize transcript accumulation in transgenic or mutant plants, respectively. F-Box and Metalloprotease reference genes were used as internal controls to normalize the amount of mRNA present in each sample. * indicates that the mean of each gene is significantly different from the control (t -test, p < 0.05).
The transcript levels of all three urease-encoding genes in the mutants plants followed the predicted pattern: eu4-a and eu3-a displayed normal mRNA levels of the three genes; eu1-a and eu1-a/eu4-a presented lower levels of Eu1, but normal Eu4 and Eu5 expression levels (Figure1B). It is worth noting that although not affecting the mRNA expression levels, the eu4-a (and eu4-b) mutant produces a non-functional enzyme with a single amino acid replacement (Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.).
Ureolytic activity
Ureolytic activity in transgenic and non-transgenic plants was evaluated with cresol red pH indicator by the seed chip assay of dried samples of leaves, roots and seeds. As expected, samples containing non-transgenic tissues showed pink coloration, indicative of urea hydrolysis. On the other hand, leaf and root samples of transgenic plants showed no observable color change even after 24 h incubation at 60 °C, indicating absence or drastic reduction of urease activity. Slices of mature seeds exhibited little or no activity. Very low urease activity was confirmed comparing transgenic with eu3-a seeds (used as a negative control) (Supplemental Figure S1).
Taken together, urease expression and activity assays indicate that transgenic plants have had all three urease-encoding genes silenced in all tissues, reinforcing the potential of these plants for functional studies.
Germination
The germination rate of T2 seeds from the two independent transgenic events was evaluated and compared to that of non-transgenic seeds. No differences were detected, suggesting that the absence of all three ureases did not affect germination. The same result was observed on germination of the eu1-a, eu4-a, eu1-a/eu4-a and eu3-a mutants and non-mutant Williams82. Germination rates were higher than 90% for all genotypes (data not shown).
Developmental pattern
Plant development was evaluated 7, 14, 21 and 30 days after sowing. T2 transgenic plants and non-transgenic plants, as well as eu1-a, eu4-a, eu1-a/eu4-a and eu3-a mutant and non-mutant plants were classified into developmental categories. Interaction among genotype, developmental categories and time-course was highly significant (p < 0.01). The two independent transgenic events showed a significant delay in development when compared with non-transgenic plants (Figure 2A and Figure 3). Size and dry weight of shoots and roots were significantly lower in transgenic plants when compared with non-transgenic (Table 2).
Plant developmental pattern over the first month after germination. (A) Transgenic and non-transgenic plants were evaluated: 18 plants derived from each event (A3 and A8) and 10 non-transgenic plants. (B) eu4-a, eu1-a, eu1-a/eu4-a and eu3-a mutant and non-mutant plants were evaluated: 18 plants from each mutant and 10 non-mutant plants. Plants were classified according categories: VE= emergence of cotyledons; VC= completely opened cotyledons; V1 = completely developed unifoliate leaf pair; V2= completely developed first trifoliate leaf; V3= completely developed second trifoliate leaf. A generalized linear model for repeated measures was used to compare the plant development among genotypes (p < 0.01).
Transgenic and non-transgenic plants seven, 14, 21 and 30 days after germination. Transgenic plants and non-transgenic plants are from cv. IAS5.
The developmental pattern of mutant plants was compared to non-mutants. The eu1-a single mutant showed a pattern similar to that of the non-mutant. The eu4-a mutant developed faster than control plants. Slower development was observed for the eu1-a/eu4-a double mutant and the eu3-a single mutant when compared to the non-mutant plants (Figure 2B). One month after germination, no differences were observed among the shoot and root sizes of eu1-a and eu4-a mutants and non-mutant plants. The shoot sizes of eu1-a/eu4-a double mutants and eu3-a single mutants were significantly smaller than those of the other genotypes. Regarding shoot and root weight, the eu1-a/eu4-a and eu3-a mutants were lighter than the other two mutants, but did not differ significantly from control. Total root length analysis showed that mutant eu3-a had the smallest root system (Table 2).
Nitrogen content
The nitrogen content was measured in shoots of one-month-old plants. The N content in transgenic plants of the two independent events was significantly lower than that present in non-transgenic plants (Figure 4A). The comparison among mutants showed that eu3-a presented the lowest and eu1-a the highest N content (Figure 4B)
Nitrogen content rate in shoot of soybean at one month after germination. (A) Transgenic and non-transgenic plants were evaluated: 18 plants derived from each event (A3 and A8) and 10 non-transgenic plants. (B) eu4-a, eu1-a, eu1-a/eu4-a and eu3-a mutant and non-mutant plants were evaluated: 18 plants from each mutant and 10 non-mutant plants. ANOVA, p < 0.0001. Means followed by the same letter did not differ by Tukey's post hoc test.
Grain yield
The number of seeds produced by three generations of transgenic plants was compared with those produced by non-transgenic plants. A significantly lower number of seeds was obtained for transgenic plants (Figure 5).
Relative percentage of seeds produced by transgenic plants in three generations (T1, T2 and T3). The mean number of seeds produced by non-transgenic plant was considered 100%. Transgenic plants and non-transgenic plants are from cv. IAS5. * indicates that the mean of transgenic seeds is significantly different from non-transgenic in each generation (t-test, p < 0.05).
Discussion
The present study aimed to evaluate the contribution of soybean ureases to seed germination and plant development. Analyses were performed using co-suppressed transgenic plants and plants with mutations in urease-related genes. The transgenic plants were the progeny (T2) of two independent events in which the Eu4 gene was down-regulated as previously described (Wiebke-Strohm et al., 2012Wiebke-Strohm B, Pasquali G, Margis-Pinheiro M, Bencke M, Bücker-Neto L, Becker-Ritt AB, Martinelli AHS, Rechenmacher C, Polacco JC, Stolf R, et al. (2012) Ubiquitous urease affects soybean susceptibility to fungi. Plant Mol Biol 78:1-13.). Molecular analyses showed that Eu4 co-suppression was maintained in the transgenic progeny. In addition it was verified that transgenic plants exhibited very low transcript levels of the other two ureases encoded by the Eu1 and Eu5 genes. The phenomenon of co-suppression by transgenic DNA has been observed in many organisms, with the introduction of transgenic copies of a gene resulting in reduced expression of the transgene, as well as of the endogenous gene. This effect depends on the sequence identity between transgene and endogenous gene (Ketting and Plasterk, 2000Ketting, RF and Plasterk, RH (2000) A genetic link between co-suppression and RNA interference in C. elegans. Nature 404:296-298.).
Soybean plants with mutations in urease genes were also evaluated confirming the expected expression pattern. Normal transcript levels of all three urease-encoding genes were detected for eu4-a and eu3-a mutants. This result is consistent with the eu4-a allele encoding G468E missense-altered ubiquitous urease (Goldraij et al., 2003Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.). Eu3 is the only UreG-encoding gene in the soybean genome. UreG is essential for urease activation, and the eu3-a mutant presents a complete loss of urease activity (Freyermuth et al., 2000Freyermuth SK, Bacanamwo M and Polacco JC (2000) The soybean Eu3-a gene encodes an Ni-binding protein necessary for urease activity. Plant J 21:53-60.), consistent with a > 90% deletion of the UreG ORF (Tezotto et al., 2016Tezotto T, Souza SCR, Mihail J, Favarin JL, Mazzafera P, Bilyeu K and Polacco JC (2016) Deletion of the single UreG urease activation gene in soybean NIL lines: Characterization and pleiotropic effects. Theor Exp Plant Physiol 28:307-320.). The eu3-a mutation did not alter Eu4, Eu1 and Eu5 expression levels, indicating a lack of feedback control on urease structural gene transcription by apo-urease(s). The mutants eu1-a and eu1-a/eu4-a exhibited similar expression patterns: lower Eu1, but normal Eu4 and Eu5 transcript levels. Since eu1-a is a null mutant (Polacco and Holland, 1993Polacco JC and Holland MA (1993) Roles of urease in plant cells. Int Rev Cytol 47:1189-1195.), low levels of Eu1 transcripts are expected, and indeed, Torisky et al. (1994)Torisky RS, Griffin JD, Yenofsky RL and Polacco JC (1994) A single gene (Eu4) encodes the tissue-ubiquitous urease of soybean. Mol Gen Genet 242:404-414. employing Eu1-specific PCR primers, recovered no detectable product from eu1-a embryo cDNA.
Ureolytic activity in transgenic and non-transgenic plants was evaluated by the seed chip assay. As expected, leaf and root samples of transgenic plants showed no color change even after 24 h incubation. The urease activity level can be inferred based on the time change “yellow to pink” on the seed chip assay (Polacco et al., 2011Polacco JC, Hyten DL, Medeiros-Silva M, Sleper DA and Bilyeu KD (2011) Mutational analysis of the major soybean UreF paralogue involved in urease activation. J Exp Bot 62:3599-3608.). According to the authors, 0.2% normal urease specific activity requires 10 hours for changing the solution color. When the activity decreases to 0.15%, the time required for solution color change is 48 hours. Based on these data, we conclude that the urease activity in leaves and roots of transgenic plants was absent or less than 0.2%, since no change in color was observed after a 24 hours incubation. The reaction catalyzed by urease is essential to allow most organisms (those lacking urea carboxylase) to use external or internally generated urea as a nitrogen source (Mobley and Hausinger, 1989Mobley HL and Hausinger RP (1989) Microbial ureases: Significance, regulation and molecular characterization. Microbiol Rev 53:85-108.; Mobley et al., 1995Mobley HL, Island MD and Hausinger RP (1995) Molecular biology of microbial ureases. Microbiol Rev 59:451-480.).
It has been demonstrated that aged A. thaliana seeds fail to germinate when urease was chemically inhibited, but seed viability could be rescued by an external N source (Zonia et al., 1995Zonia LE, Stebbins N and Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107:1097-1103.). In the present study no differences were detected in germination rates of transgenic and mutant seeds. However, it is important to highlight that soybean seeds were not aged and have a much higher protein content than A. thaliana seeds.
An association between urease activity and developmental pattern was observed. Transgenic plants, as well as eu3-a and the double eu1-a/eu4-a mutants, showed a delay over the first month after sowing. The delay in development was maintained even in adult transgenic plants and may be the cause of lower seed production. In A. thaliana, both urease transcripts and ureolytic activity increased after germination, especially in 8/9-day-old wild-type seedlings (Zonia et al., 1995Zonia LE, Stebbins N and Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107:1097-1103.). Embryo-specific urease (Eu1) activity in young soybean plants was also observed by Torisky and Polacco, (1990)Torisky RS and Polacco JC (1990) Soybean roots retain the seed urease isozyme synthesized during embryo development. Plant Physiol 94:681-689.. Similarly, high transcript level of Eu4 and moderate transcript levels of Eu1 and Eu5 were detected in soybean seeds one day after dormancy break (Wiebke-Strohm et al., 2016Wiebke-Strohm B, Ligabue-Braun R, Rechenmacher C, de Oliveira-Busatto LA, Carlini CR and Bodanese-Zanettini MH (2016) Structural and transcriptional characterization of a novel member of the soybean urease gene family. Plant Physiol Biochem 101:96-104.). Taken together, these results indicate that increased urease content and/or ureolytic activity play a role in early stages of plant development.
A role in making nitrogen available during plant development has been attributed to soybean ubiquitous urease due to its catalytic activity and tissue distribution (Stebbins et al., 1991Stebbins N, Holland MA, Cianzio SR and Polacco JC (1991) Genetic tests of the roles of the embryonic ureases of soybean. Plant Physiol 97:1004-1010.). However, our results indicate that Eu1 and Eu5 have a contribution in the developmental process as well. This is supported by the finding that transgenic A3 and A8, mutant eu1-a/eu4-a and eu3-a plants showed a delay in the first developmental stages. In transgenic plants, the impairment in development was confirmed by the significant reduction in size and weight of roots and shoots one month after germination. These data are consistent with significantly lower nitrogen content detected in transgenic plants. Regarding mutant plants, eu3-a tends to be smaller and lighter than the other genotypes, although significant differences were only detected for roots and shoots size. The reduction in dry matter is also reflected in the lowest nitrogen content. An unexpected result was the significantly higher nitrogen content present in eu1-a. The differences in nitrogen content might be due to differences in mutant genetic backgrounds. Williams is the background for eu1-a, while Williams82 is the one for the other mutants. In our experiment, non-mutant Williams82 was used as control.
Based on bioinformatics analyses, Eu5 has been suggested not to be a functional ureolytic enzyme due to a number of mutations, including deletions (Witte, 2011Witte CP (2011) Urea metabolism in plants. Plant Sci 180:431-438.). However, according to our results the product of this gene might be involved in plant development. Further studies are necessary to elucidate whether the ureolytic activity and/or other non-enzymatic property of ureases are involved on plant development.
Acknowledgments
We thank Dr. Stela Maris de Jezus Castro (UFRGS) for statistical analysis; Dr Christian Bredemeier for supervision on nitrogen analysis; the CCGL-Tec research team and Silvia Richter for technical assistance. This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS).
References
- Bernard RL and Cremeens CR (1988) Registration of Williams 82 soybean. Crop Sci 28:1027-1028.
- Bohner A, Kojima S, Hajirezaei M, Melzer M and Von Wiren N (2015) Urea retranslocation from senescing Arabidopsis leaves is promoted by DUR3-mediated urea retrieval from leaf apoplast. Plant J 81:377-387.
- Cao F-Q, Werner AK, Dahncke K, Romeis T, Liu L-H and Witte CP (2010) Identification and characterization of proteins involved in rice urea and arginine catabolism. Plant Physiol 15:98-108.
- Carlini C and Ligabue-Braun R (2016) Ureases as multifunctional toxic proteins: A review. Toxicon 110:90-109.
- Carter EL, Flugga N, Boer JL, Mulrooney SB and Hausinger RP (2009) Interplay of metal ions and urease. Metallomics 1:207-221.
- Dixon NE, Gazzola TC, Blakeley RL and Zermer B (1975) Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J Am Chem 97:4131-4133.
- Eskew DL, Welch RM and Cary EE (1983) Nickel: An essential micronutrient for legumes and possibly all higher plants. Science 222:621-623.
- Freyermuth SK, Bacanamwo M and Polacco JC (2000) The soybean Eu3-a gene encodes an Ni-binding protein necessary for urease activity. Plant J 21:53-60.
- Goldraij A and Polacco JC (1999) Arginase is inoperative in developing soybean seeds. Plant Physiol 119:297-304.
- Goldraij A and Polacco JC (2000) Arginine degradation by arginase in mitochondria of soybean seedling cotyledons. Planta 210:652-658.
- Goldraij A, Beamer LJ and Polacco JC (2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132:1801-1810.
- Hogan ME, Swift IE and Done J (1983) Urease assay and ammonia release from leaf tissues. Phytochemistry 22:663-700.
- Jian B, Liu B, Bi Y, Hou W, Wu C and Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59.
- Kanamori T, Kanou N, Atomi H and Imanaka T (2004) Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol 186:2532-2539.
- Ketting, RF and Plasterk, RH (2000) A genetic link between co-suppression and RNA interference in C. elegans. Nature 404:296-298.
- Krajewska B (2009) Ureases I. Functional, catalytic and kinetic properties: A review. J Mol Catal B Enzymatic 59:9-21.
- Libault MS, Thibivilliers DD, Bilgin O, Radwan M, Benitez SJ, Clough and Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1:44-54.
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408.
- Marschner P (2012) Marschner's Mineral Nutrition of Higher Plants. 3rd edition. Academic Press, London, 651p.
- Meyer-Bothling LE and Polacco JC (1987) Mutational analysis of the embryo-specific urease locus of soybean. Mol Gen Genet 209:439-444.
- Micallef BJ and Shelp BJ (1989) Arginine metabolism in developing soybean cotyledons. I. Relationship to nitrogen nutrition. Plant Physiol 90:624-630.
- Mobley HL and Hausinger RP (1989) Microbial ureases: Significance, regulation and molecular characterization. Microbiol Rev 53:85-108.
- Mobley HL, Island MD and Hausinger RP (1995) Molecular biology of microbial ureases. Microbiol Rev 59:451-480.
- Neumaier N, Nepomuceno AL and Farias JRB (2000) Estádios de desenvolvimento da cultura de soja. In: Bonato ER (ed). Estresses em Soja. EMBRAPA CNPT, Passo Fundo, pp 19-44.
- Polacco JC and Havir EA (1979) Comparisons of soybean urease isolated from seed and tissue culture. J Biol Chem 254:1707-1715.
- Polacco JC and Holland MA (1993) Roles of urease in plant cells. Int Rev Cytol 47:1189-1195.
- Polacco JC and Winkler RG (1984) Soybean leaf urease: A seed enzyme? Plant Physiol 74:800-803.
- Polacco JC, Krueger RW and Winkler RG (1985) Structure and possible ureide degrading function of the ubiquitous urease of soybean. Plant Physiol 79:794-800.
- Polacco JC, Hyten DL, Medeiros-Silva M, Sleper DA and Bilyeu KD (2011) Mutational analysis of the major soybean UreF paralogue involved in urease activation. J Exp Bot 62:3599-3608.
- Polacco JC, Mazzafera P and Tezotto T (2013) Opinion - Nickel and urease in plants: Still many knowledge gaps. Plant Science 199-200:79-90.
- Sirko A and Brodzik R (2000) Plant ureases: Roles and regulation. Acta Biochim Pol 47:1189-1195.
- Stebbins N and Polacco JC (1995) Urease is not essential for ureide degradation in soybean. Plant Physiol 109:169-175.
- Stebbins N, Holland MA, Cianzio SR and Polacco JC (1991) Genetic tests of the roles of the embryonic ureases of soybean. Plant Physiol 97:1004-1010.
- Tedesco MJ, Gianello C, Bissani CA, Bohnen H and Vokweiss SJ (1995) Análise de Solo, Plantas e Outros Materiais. Boletim Técnico, 5. 3rd edition. UFRGS, Porto Alegre, 174 p.
- Tezotto T, Souza SCR, Mihail J, Favarin JL, Mazzafera P, Bilyeu K and Polacco JC (2016) Deletion of the single UreG urease activation gene in soybean NIL lines: Characterization and pleiotropic effects. Theor Exp Plant Physiol 28:307-320.
- Torisky RS and Polacco JC (1990) Soybean roots retain the seed urease isozyme synthesized during embryo development. Plant Physiol 94:681-689.
- Torisky RS, Griffin JD, Yenofsky RL and Polacco JC (1994) A single gene (Eu4) encodes the tissue-ubiquitous urease of soybean. Mol Gen Genet 242:404-414.
- Van Etten CH, Miller RW, Wulf IA and Jones Q (1963) Amino acid composition of seeds from 200 angiospermous plant species. J Agric Food Chem 11:399-410.
- Wang WH, Kohler B, Cao FQ and Liu LH (2008) Molecular and physiological aspects of urea transport in higher plants. Plant Sci 175:467-477.
- Wiebke-Strohm B, Pasquali G, Margis-Pinheiro M, Bencke M, Bücker-Neto L, Becker-Ritt AB, Martinelli AHS, Rechenmacher C, Polacco JC, Stolf R, et al (2012) Ubiquitous urease affects soybean susceptibility to fungi. Plant Mol Biol 78:1-13.
- Wiebke-Strohm B, Ligabue-Braun R, Rechenmacher C, de Oliveira-Busatto LA, Carlini CR and Bodanese-Zanettini MH (2016) Structural and transcriptional characterization of a novel member of the soybean urease gene family. Plant Physiol Biochem 101:96-104.
- Witte CP (2011) Urea metabolism in plants. Plant Sci 180:431-438.
- Witte CP, Tiller SA, Taylor MA and Davies HV (2002) Leaf urea metabolism in potato. Urease activity profile and patterns of recovery and distribution of (15) N after foliar urea application in wild-type and urease-antisense transgenics. Plant Physiol 128:1129-1136.
- Zonia LE, Stebbins N and Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107:1097-1103.
-
Associate Editor: Santiago Mora-Garcia
Supplementary Material
The following online material is available for this article:
Figure S1 - Ureolytic activity in transgenic and non-transgenic plants.
Publication Dates
-
Publication in this collection
02 Mar 2017 -
Date of issue
2017
History
-
Received
13 Apr 2016 -
Accepted
10 Aug 2016