Abstract
Alstroemeria L. (Alstroemeriaceae) represents one of the most diverse genera of vascular plants in Chile. It contains approximately 54 taxa, 40 of which are endemic. The “complex” Alstroemeria magnifica is endemic to Chile, and it comprises four varieties: A. magnifica var. magenta, A. magnifica var. magnifica, A. magnifica var. sierrae, and A. magnifica var. tofoensis. It is distributed from Coquimbo to the Valparaíso Region. We analyzed karyotypes of 10 populations along its natural distribution. All the populations presented an asymmetric karyotype, with 2n = 16 chromosomes but with three different karyotypic formulae. Alstroemeria magnifica var. magnifica and A. magnifica var. sierrae presented the same karyotypic fomula, and A. magnifica var. magenta, and A. magnifica var. tofoensis each had a different formula. The scatter plot among CVCL vs. MCA shows different groupings between populations of the four varieties. Based on the results, it is possible to consider raising Alstroemeria magnifica var. magenta to species level (A. magenta) and A. magnifica var. tofoensis to subspecies level (A. magnifica subsp. tofoensis); A. magnifica var. magnifica and A. magnifica var. sierrae should each remain as varieties. Nevertheless, these taxonomic changes should be considered tentative, as additional sources of evidence become available.
Keywords:
Alstroemeria; karyotype; species complex; asymmetry; Chile
Introduction
Alstroemeria is a South American genus, which comprises about 82 taxa distributed from Venezuela (3°N) to Tierra del Fuego (53°S) (Muñoz and Moreira, 2003Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p.). The centers of distribution of this genus are located in Central Chile and East of Brazil, representing a disjoint pattern of distribution produced by the isolating effect of the Cordillera de Los Andes and the South American Arid Diagonal (Muñoz and Moreira, 2003Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p.; Hofreiter, 2007Hofreiter A (2007) Biogeography and ecology of the Alstroemeriaceae–Luzuriagaceae clade in the high-mountain regions of Central and South America. Harv Pap Bot 12:259-284.; Chacón et al., 2012aChacón J, De Assis M, Meerow A and Renner S (2012a) From east Gondwana to Central America: Historical biogeography of the Alstroemeriaceae. J Biogeogr 39:1806-1818.).
In Chile, Alstroemeria represents one of the most diverse genera of vascular plants, comprising 49 taxa (33 species, 8 varieties, and 8 subspecies); 40 of which are endemic (Muñoz and Moreira, 2003Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p.). Recent studies suggest increasing to 54 the number of taxa recognized in Alstroemeria, with the validation of Alstroemeria citrina Phil. (Eyzaguirre, 2008Eyzaguirre M (2008) Validación de Alstroemeria citrina Phil. (Alstroemeriaceae). Gayana Bot 65:241-244.) and Alstroemeria parvula Phil. (Muñoz et al., 2011Muñoz M, Morales V and Moreira A (2011) Validación de Alstroemeria parvula Phil. (Alstroemeriaceae). Gayana Bot 68:114-116.). These modifications also include the discovery of Alstroemeria hookeri Lodd. subsp. sansebastiana C.M. Baeza & E. Ruiz (Baeza and Ruiz, 2011Baeza C and Ruiz E (2011) Alstroemeria hookeri Lodd. subsp. sansebastiana C.M. Baeza & E. Ruiz, nueva para la flora de Chile. Gayana Bot 68:313-315.), Alstroemeria marticorenae Negritto & C. M. Baeza (Negritto et al., 2015Negritto M, Baeza C, Ruiz E and Novoa P (2015) Alstroemeria marticorenae (Alstroemeriaceae), a new species from central Chile. Syst Bot 40:69-74.) and Alstroemeria traudliae (Hoffmann et al., 2015Hoffmann A, Watson J and Flores A (2015) Flora silvestre de Chile. Cuando el desierto florece. Volumen 1. Monocotiledóneas y otros taxones. Fundación Claudio Gay. Santiago, Salesianos impresores S.A. 261 p.).
Reports of chromosome studies in Alstroemeria are dated since 1882, recognizing a fundamental karyotype on about 30 taxa, 22 of them from Chilean species (Chacón et al., 2012bChacón J, Sousa A, Baeza C and Renner S (2012b) Ribosomal DNA distribution and a genus-wide phylogeny reveal patterns of chromosomal evolution in Alstroemeria (Alstroemeriaceae). Am J Bot 99:1501-1512.). A stable chromosome set of 2n = 16 was determined, with an asymmetric and bimodal karyotype of eight chromosomes: three or four are acrocentric and four or five are metacentric, submetacentric or subtelocentric (Baeza et al., 2008Baeza C, Schrader O, Ruiz E and Negritto M (2008) Alstroemeria presliana Herb. (Alstroemeriaceae) in Chile from a cytogenetic perspective. Chilean J Agric Res 68:328-333.). Until today, no reports of polyploids have been observed in natural populations of Alstroemeria (Baeza et al., 2007aBaeza C, Schrader O, Ruiz E and Negritto M (2007a) Análisis comparativo del cariotipo en poblaciones de Alstroemeria aurea R. Graham (Alstroemeriaceae) de Chile. Gayana Bot 64:33-39.).
Cytogenetic studies have proven useful for the delimitation of entities in Alstroemeria since every studied taxon presents a distinctive, unique, and largely stable karyotype. As such, these studies have contributed not only to the delimitation of species and varieties, but also to elicit underlying processes - at chromosomal levels - that determine the divergence of these taxa (Baeza et al., 2007bBaeza C, Schrader O and Budahn H (2007b) Characterization of geographically isolated accessions in five Alstroemeria L. species using FISH of tandemly repeated DNA sequences and RAPD analysis. Plant Syst Evol 269: 1-14.). In taxonomic complexes, a clear-cut discrimination of intraspecific taxa has resulted from the study of differences in the architecture and/or the asymmetry of chromosomes (Cajas et al., 2009Cajas D, Baeza M, Ruiz E and Negritto M (2009) Análisis citogenético en poblaciones de Alstroemeria hookeri Lodd. subsp. hookeri (Alstroemeriaceae) en la Región del Bío-Bío, Chile. Gayana Bot 66:117-126.; Baeza et al., 2010Baeza C, Ruiz E and Negritto M (2010) Comparative karyotypic analysis in the Alstroemeria hookeri Lodd. (Alstroemeriaceae) complex sensu Bayer (1987). Genet Mol Biol 33:119-124.). For example, the study of karyotype was determinant for the delimitation of taxa within the Alstroemeria hookeri complex, providing evidence for the existence of Alstroemeria hookeri subsp. sansebastiana (Baeza and Ruiz, 2011Baeza C and Ruiz E (2011) Alstroemeria hookeri Lodd. subsp. sansebastiana C.M. Baeza & E. Ruiz, nueva para la flora de Chile. Gayana Bot 68:313-315.) and supporting the proposal of Muñoz and Moreira (2003)Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p. to raise the status of Alstroemeria hookeri subsp. cummingiana to species level.
The difficult differentiation and delimitation of taxa within Alstroemeria have led to the definition of intraspecific complexes, which consist of two or four subspecies and/or varieties of the same species. Some of these complexes have become relevant taxa for the prospection of development in different areas of national interest, given their economic potential as ornamental plants, and/or their importance as representative taxa of the Chilean biodiversity. Traditional taxonomic treatments in Alstroemeria have largely been based on patterns of variability of their conspicuous flowers, which present tepals with a wide display of morphological and coloring patterns (Bayer, 1987Bayer E (1987) Die Gattung Alstroemeria in Chile. Mitt Bot Staatssamml Muenchen 24:1-362.; Muñoz and Moreira, 2003Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p.). These structures, while useful for discrimination at interspecific levels, usually exhibit levels of variation beyond levels of stability required for a robust taxonomic discrimination in several groups of species (Baeza et al., 2010Baeza C, Ruiz E and Negritto M (2010) Comparative karyotypic analysis in the Alstroemeria hookeri Lodd. (Alstroemeriaceae) complex sensu Bayer (1987). Genet Mol Biol 33:119-124., 2015Baeza C, Finot L and Ruiz E (2015) Comparative karyotype analysis of population in the Alstroemeria presliana Herbert (Alstroemeriaceae) complex in Chile. Genet Mol Biol 38:199-104., 2016aBaeza C, Finot V, Ruiz E, Carrasco P, Novoa P and Rosas M (2016a) Análisis citotaxonómico del complejo Alstroemeria diluta E. Bayer (Alstroemeriaceae) de Chile. Gayana Bot 73:221-227.,bBaeza C, Finot V, Ruiz E, Carrasco P, Novoa P, Stuessy T and González A (2016b) Comparative karyotypic analysis and cytotaxonomy in the Alstroemeria ligtu L. (Alstroemeriaceae) complex of Chile. Braz J Bot 39:305-313.). Therefore, since any potential development in these groups has been mostly restricted to inconclusive taxonomic interpretations the use of other possible sources of evidence, like cytological characters, could result useful to help taxonomic discrimination at intraspecific levels.
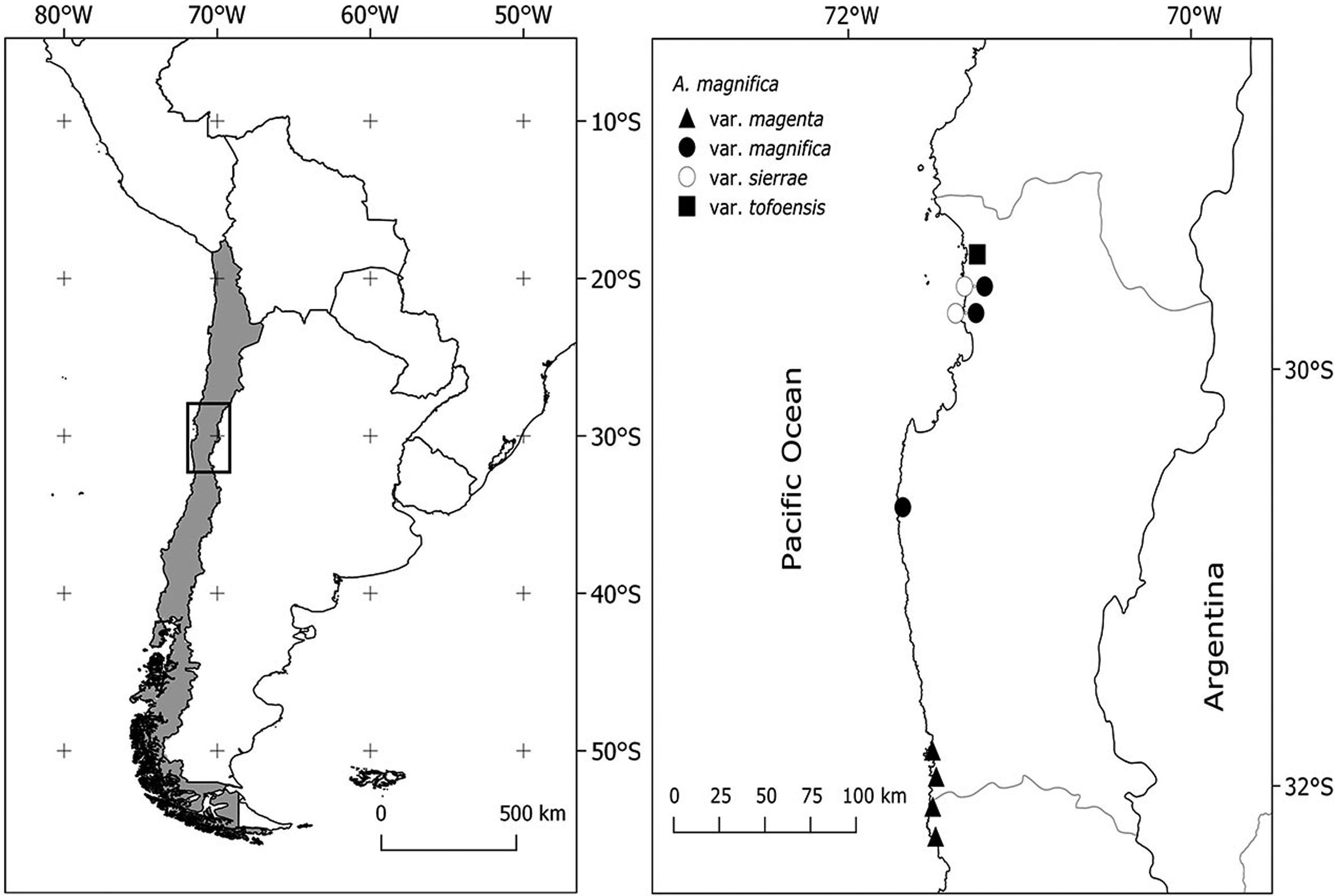
Alstroemeria magnifica Herb. is a species complex which comprises four varieties (Muñoz, 2003Muñoz M (2003) Notas nomenclaturales sobre Alstroemeria L. (Alstroemeriaceae) Not Mens Mus Hist Nat 352: 22.; Muñoz and Moreira, 2003Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p.). Along with A. hookeri, A. magnifica is one of the richest in terms of number of taxa in Alstroemeria. This complex is distributed from the locality of Chungungo (29°26’S; Region of Coquimbo) to the north of Papudo (32°21’S; Region of Valparaiso). It is recurrent across coastal rocky bluffs and slopes, most likely in areas with permanent fog. The specific ranges of distribution and description of floral morphology are the following:
Alstroemeria magnifica Herb. var. magnifica: Distributed from 29º30’S to 30º 50’S. It is recognized by their whitish to lilaceous flowers, with a plain internal tepal (no design present; Figure 1A).
Photographic representation of varieties present in the A. magnifica complex. (A) Photography of A. magnifica var. magnifica; (B) Photography of A. magnifica var. magenta; (C) Photography of A. magnifica var. sierrae; (D) Photography of A. magnifica var. tofoensis. Bar = 2 cm.
Alstroemeria magnifica Herb. var. magenta (Ehr.Bayer) Muñoz-Schick: This variety presents the largest range of distribution within the complex, ranging from 30°39’S to 32°21’S. It is distinguished by the presence of both small inflorescences and flowers. Their internal upper tepals show thick lines, which end in a respective spot at the apex. The internal lower tepal can present design or not (Figure 1B).
Alstroemeria magnifica Herb. var. sierrae (Muñoz) Muñoz-Schick: This variety occurs in a restricted distribution, from 29º36’S to 29°45’S. It presents flowers of large size, which are distinctive by the design present in the internal upper tepals, and having lines forming a large spot at the apex and basis of this structure. The internal lower tepal can present design (Figure 1C).
Alstroemeria magnifica Herb. var. tofoensis Muñoz-Schick: This variety has a very restricted distribution, from 29°26’S to 29°32’S. It is characterized by their internal upper tepals with a patch of a yellowish spot and a white background, which does not reach the borders. Scattered lines with no spotty end at the apex are also present. The internal inferior tepal is maculate at the basis (Figure 1D).
Until now, cytological work has not been extensive for completely clarifying the taxonomic status of the infraspecific taxa of the A. magnifica complex. Buitendijk and Ramanna (1996)Buitendijk J and Ramanna M (1996) Giemsa C-banded karyotypes of eigth species of Alstroemeria L. and some of their hybrids. Ann Bot 78:449-457. analyzed the karyotype of A. magnifica subsp. magnifica, informing the morphology of chromosomes and interspecific variability in the C-bands patterns. Additional reports exist about the genomic size (Buitendijk et al., 1997Buitendijk J, Boon E and Ramanna M (1997) Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Ann Bot 79:343-353., 1998Buitendijk J, Peters A, Jan-Quené R and Ramanna M (1998) Genome size variation and C-band polymorphism in Alstroemeria aurea, A. ligtu and A. magnifica (Alstroemeriaceae). Plant Syst Evol 212:87-106.), which in association with patterns of variation in C-bands, suggest discontinuous variation in the quantity of nuclear DNA of A. magnifica. Nevertheless, since these observations are circumscribed to cultivated specimens only (mostly cultivars and greenhouse varieties), no additional cytological information exists from local populations and/or range of variation within their natural environmental and geographic ranges.
Therefore, the present study aims to characterize and compare, at a cytotaxonomic level, the four varieties of the A. magnifica complex. Thus, using representative sampling from the total of the geographic range of distribution, we expect to offer a suggested clarification of the taxonomic status of each variety within the complex.
Materials and Methods
Plant material
A total of two to four individuals from 10 populations of A. magnifica were collected across the known range of distribution (Table 1). Voucher specimens from each population were deposited in the Herbarium of the University of Concepción (CONC). Figure 2 shows the distribution of the collected populations, which were used in the present study.
Methodology for the study of karyotypes
Rhizome roots (1-2 cm length) obtained from individuals in each population and held in a greenhouse, were cut and pre-treated with a solution of 8-hydroxyquinoline (2 mM) for 24 h at 4 ºC. These samples were subsequently fixed with a fresh solution of ethanol/acetic acid (3:1) for 24 h. Squash preparations from root tips were made using an acid hydrolysis pretreatment with HCL 0.5 N during 17 min at 42 ºC. After washing in distilled water, the material was stained with 1% orcein solution. Metaphase chromosome plates were photographed using a Zeiss Axioskop hmicroscope, with an incorporated video camera. Chromosomes were measured with the assistance of the software MicroMeasure 3.3 (Reeves, 2001Reeves A (2001) MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 44:239-443.) and classified according to arm ratios (long arm/short arm; modified from Levan et al., 1964Levan A, Fredga K and Sandberg A (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201-220.). From 10 metaphase plates in each analyzed population, randomly chosen from the total of individuals, an idiogram was constructed for each studied variety. Intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) indices were calculated for each analyzed population, following the proposal of Peruzzi and Erôglu (2013)Peruzzi L and Erôglu H (2013) Karyotype asymmetry: again, how to measure and what to measure? CompCytogen 7:1-9.. Both indices were placed in a scatter plot, accompanied by the total length of diploid chromosomes (TCL), for each analyzed population using the package plot3D v 1.1 (Soetaert, 2016Soetaert K (2016) plot3D: Plotting Multi-Dimensional Data. R package version 1.1. https://CRAN.R-project.org/package=plot3D.
https://CRAN.R-project.org/package=plot3...
) in R v 3.3.3 (R Core Team, 2017R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
https://www.R-project.org/...
).
Results and Discussion
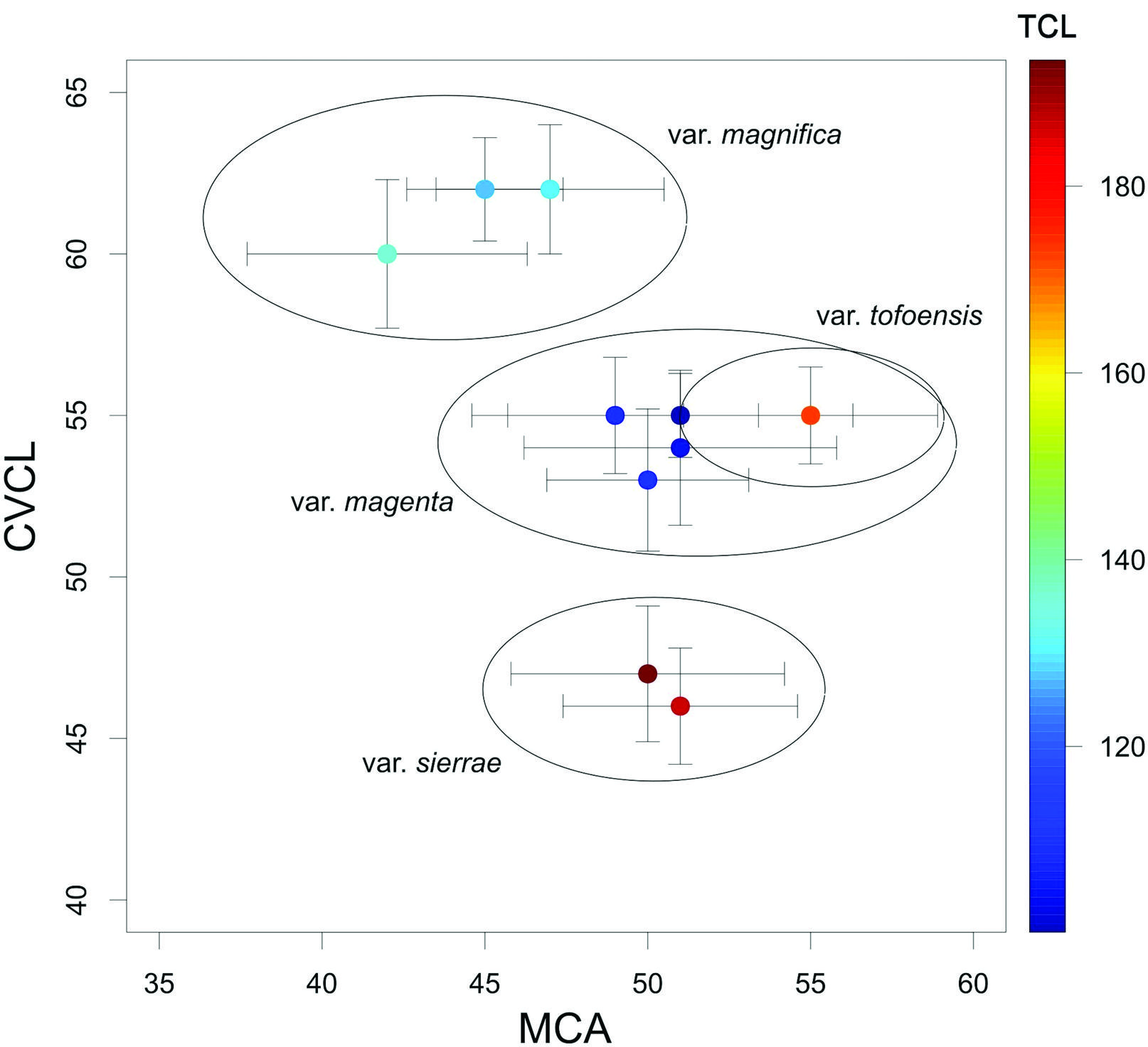
All analyzed populations of A. magnifica revealed a 2n = 2x = 16. A. magnifica var. magnifica and A. magnifica var. sierrae presented the same haploid formula: two pairs of metacentric chromosomes, one submetacentric pair, two subtelocentric pairs, two subtelocentric pairs with satellite and one telocentric pair with satellite (2m + 1sm + 2st + 2st-sat + 1t-sat; Figure 3A,C). A. magnifica var. magenta presented a haploid formula of two metacentric chromosomes, two submetacentric pairs, one subtelocentric pair, two subtelocentric pairs with satellite and one telocentric pair with satellite (2m + 2sm +1st + 2st-sat + 1t-sat; Figure 3B). A. magnifica var. tofoensis presented a haploid formula of two metacentric chromosomes, one submetacentric pair, one subtelocentric pair, two telocentric pairs and two telocentric pairs with satellite (2m + 1sm + 1st + 2t + 2t-sat; Figure 3D). Figure 4 shows representative metaphase plates for each studied taxon. The values of CVCL, MCA and TCL per populations are summarized in Table 2. Figure 5 represents the scatter plot of CVCL and MCA indices.
Idiograms of A. magnifica varieties. (A) A. magnifica var. magnifica; (B) A. magnifica var. magenta; (C) A. magnifica var. sierrae; (D) A. magnifica var. tofoensis.
Metaphase plates in the varieties of A. magnifica complex. (A) A. magnifica var. magnifica (4408); (B) A. magnifica var. sierrae (4406); (C) A. magnifica var. magenta (4381); (D) A. magnifica var. tofoensis (4409). Bar = 5 μm.
Karyotype features of the varieties of Alstroemeria magnifica. CVCL = Coefficient of variation of chromosome length; MCA = Mean centromeric asymmetry index according to Peruzzi and Erôglu (2013)Peruzzi L and Erôglu H (2013) Karyotype asymmetry: again, how to measure and what to measure? CompCytogen 7:1-9.; SD = Standard deviation; TLC = Total length in diploid chromosomes.
Scatter plot among populations of Alstroemeria magnifica varieties using values of MCA vs. CVCL.
Karyological studies have previously reported about the morphology of chromosomes, patterns and polymorphism of C-bands, nuclear content, and the genomic size of A. magnifica (Buitendijk and Ramanna, 1996Buitendijk J and Ramanna M (1996) Giemsa C-banded karyotypes of eigth species of Alstroemeria L. and some of their hybrids. Ann Bot 78:449-457.; Buitendijk et al., 1997Buitendijk J, Boon E and Ramanna M (1997) Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Ann Bot 79:343-353., 1998Buitendijk J, Peters A, Jan-Quené R and Ramanna M (1998) Genome size variation and C-band polymorphism in Alstroemeria aurea, A. ligtu and A. magnifica (Alstroemeriaceae). Plant Syst Evol 212:87-106.). The present study concurs with the findings made in those publications, specifically on the stability of the 2n = 16 present in all varieties of A. magnifica. Additionally, our results support the typical asymmetric and bimodal karyotype present in Alstroemeria, with four to seven metacentric, submetacentric or subtelocentric chromosomes (Baeza et al., 2008Baeza C, Schrader O, Ruiz E and Negritto M (2008) Alstroemeria presliana Herb. (Alstroemeriaceae) in Chile from a cytogenetic perspective. Chilean J Agric Res 68:328-333.). Within this cytological configuration, it is possible to distinguish, at least, two patterns that can discriminate the varieties of this complex.
First, while identical in structure, notorious differences are noticeable in the total length of chromosomes (TLC). This observation allows to distinguish A. magnifica var. magnifica from A. magnifica var. sierrae, as the former exhibits smaller chromosomes than the latter (Table 2), despite exhibiting identical karyotypes (Figures 3A and 3C). Populations from A. magnifca var. sierrae and A. magnifica var. magnifica are separated because of their differences in the CVCL index (Figure 5), which is directly related to the TLC values (Perruzzi and Erôglu, 2013Peruzzi L and Erôglu H (2013) Karyotype asymmetry: again, how to measure and what to measure? CompCytogen 7:1-9.). This cytological pattern is intriguing, because of its recurrent presence in other complexes in Alstroemeria. For example, in the Alstroemeria diluta complex, both recognized subspecies, A. diluta subsp. diluta and A. diluta subsp. chrysantha, reveal similar karyotypes but a different TLC values (Baeza et al., 2016aBaeza C, Finot V, Ruiz E, Carrasco P, Novoa P and Rosas M (2016a) Análisis citotaxonómico del complejo Alstroemeria diluta E. Bayer (Alstroemeriaceae) de Chile. Gayana Bot 73:221-227.). Such change in chromosome lenght could be the result of changes in the total genomic nuclear size of A. magnifica (Buitendijk et al., 1997Buitendijk J, Boon E and Ramanna M (1997) Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Ann Bot 79:343-353., 1998Buitendijk J, Peters A, Jan-Quené R and Ramanna M (1998) Genome size variation and C-band polymorphism in Alstroemeria aurea, A. ligtu and A. magnifica (Alstroemeriaceae). Plant Syst Evol 212:87-106.), which could have implications as a mechanism of differentiation among taxa in Alstroemeria. Nonetheless, this circumstantial evidence should be further corroborated with additional studies based on nuclear DNA content (e.g., flow cytometry) and its variation across natural populations.
The second pattern is exhibited in A. magnifica var. magenta and A. magnifica var. tofoensis, which present different and unique karyotypes – compared to A. magnifica var. magnifica and A. magnifica var. sierrae (Figures 3 and 4). In this case, chromosome 3 of A. magnifica var. magenta is submetacentric, instead of subtelocentric or telocentric found in the other varieties of A. magnifica. In A. magnifica var. tofoensis, a polymorphism in the length of chromosome arms is detected between homologous chromosomes of pair 5, which is also expressed in terms of greater levels of magnitude in standard variation related to TCL (Table 2). This pattern is in line with previous reports in A. philippii, where a population revealed length polymorphism between homologous in the chromosome pair 3 (Buitendijk et al., 1998Buitendijk J, Peters A, Jan-Quené R and Ramanna M (1998) Genome size variation and C-band polymorphism in Alstroemeria aurea, A. ligtu and A. magnifica (Alstroemeriaceae). Plant Syst Evol 212:87-106.). A similar situation has been found in species of Brachycome, Triticum, Tulpia, Secale, Allium (Houben et al., 2000Houben A, Wanner G, Hanson L, Verlin D, Leach C and Timmis J (2000) Cloning and characterization of polymorphic heterochromatic segments of Brachycome dichromosomatica. Chromosoma 109:206-213.), Scilla (Greilhuber and Speta, 1976Greilhuber J and Speta F (1976) C-banded karyotypes in the Scilla hohenackeri group S. persica, and Poschkina (Liliaceae). Plant Syst Evol 126:149-188.), Placea amoena (Baeza and Schrader, 2004Baeza C and Schrader O (2004) Karyotype analysisof Placea amoenaPhil. (Amaryllidaceae) by double fluorescencein situhybridization. Caryologia 57:200-2005.), and Chaetanthera pentacaenoides (Baeza and Torres-Díaz, 2006Baeza C and Torres-Díaz C (2006) El cariotipo de Chaetanthera pentacaenoides (Phil.) Hauman (Asteraceae). Gayana Bot 63:180-182.).
In terms of the overall differentiation among the varieties of the A. magnifica complex, a better characterization is possible to achieve by using individual patterns of asymmetry in chromosomes. A. magnifica var. magnifica and A. magnifica var. magenta revealed substantial differences in their MCA and CVCL values, which also occurs with A. magnifica var. tofoensis; nonetheless, the latter with similar patterns than A. magnifica var. magenta (Figure 5). Despite this, the karyotype of A. magnifica var. magnifica and A. magnifica var. magenta exhibits notorious differences, especially in the unique presence of a submetacentric chromosome in pair 3 of A. magnifica var. magenta compared to the rest of the complex. Furthermore, A. magnifica var. tofoensis presents a polymorphism in the homologous metacentric chromosomes in pair 5 (see above), while this is subtelocentric in A. magnifica var. magenta. These unique features suggest that both asymmetry patterns and karyotype variation should be considered together if this evidence is to be used for the precise discrimination of the involved varieties.
The results of this study suggest that patterns of chromosome variation can be instrumental for discriminating among taxa and proposing taxonomic rearrangements in the species complexes of Alstroemeria, as they tend to exhibit higher levels of stability and resolution than traditional tepal morphological characters at intraspecific levels (Cajas et al., 2009Cajas D, Baeza M, Ruiz E and Negritto M (2009) Análisis citogenético en poblaciones de Alstroemeria hookeri Lodd. subsp. hookeri (Alstroemeriaceae) en la Región del Bío-Bío, Chile. Gayana Bot 66:117-126.; Baeza et al., 2010Baeza C, Ruiz E and Negritto M (2010) Comparative karyotypic analysis in the Alstroemeria hookeri Lodd. (Alstroemeriaceae) complex sensu Bayer (1987). Genet Mol Biol 33:119-124., 2015Baeza C, Finot L and Ruiz E (2015) Comparative karyotype analysis of population in the Alstroemeria presliana Herbert (Alstroemeriaceae) complex in Chile. Genet Mol Biol 38:199-104., 2016aBaeza C, Finot V, Ruiz E, Carrasco P, Novoa P and Rosas M (2016a) Análisis citotaxonómico del complejo Alstroemeria diluta E. Bayer (Alstroemeriaceae) de Chile. Gayana Bot 73:221-227.,bBaeza C, Finot V, Ruiz E, Carrasco P, Novoa P, Stuessy T and González A (2016b) Comparative karyotypic analysis and cytotaxonomy in the Alstroemeria ligtu L. (Alstroemeriaceae) complex of Chile. Braz J Bot 39:305-313.). In this case, these changes would be further supported, as cytological data is integrated and contextualized with preliminary results observed from additional character sources (chloroplast DNA, colorimetric variation and morphometry of tepals; Carrasco et al. in preparation). For example, given the concordance of cytological data with patterns of discrete morphological variation, sympatric distribution and differentiation based in chloroplast DNA, it is likely that A. magnifica var. sierrae and A. magnifica var. magnifica should retain their taxonomic status without modifications. Instead, A. magnifica var. tofoensis status should be changed to subspecies level, because, despite presenting clear differentiation in floral characters, isolated distribution, and a distinctive unique karyotype, it presents close genetic similarity with A. magnifica var. magnifica and A. magnifica var. sierrae. Likewise, it would be recommendable to revalidate A. magenta Bayer, from A. magnifica var. magenta, as originally proposed by Bayer (1987)Bayer E (1987) Die Gattung Alstroemeria in Chile. Mitt Bot Staatssamml Muenchen 24:1-362., given its consistent differences in vegetative and floral characters (smaller plants and flowers), allopatric distribution, a distinctive karyotype and substantial genetic distance from the rest of the taxa of A. magnifica complex. Nonetheless, these proposals should be seen as tentative, contingent upon additional and more conclusive results that can be added from the suggested character sources.
Acknowledgments
This study was supported by FONDECYT Nº 1130349 and FONDECYT Nº 3160453. The authors thank the Corporación Nacional Forestal (CONAF) for providing the permits that allowed collecting specimens in the Fray Jorge National Park. Thanks are also to the Departamento de Botánica and the Herbario de la Universidad de Concepción (CONC) for their assistance provided in this study.
References
- Baeza C and Schrader O (2004) Karyotype analysisof Placea amoenaPhil. (Amaryllidaceae) by double fluorescencein situhybridization. Caryologia 57:200-2005.
- Baeza C and Torres-Díaz C (2006) El cariotipo de Chaetanthera pentacaenoides (Phil.) Hauman (Asteraceae). Gayana Bot 63:180-182.
- Baeza C and Ruiz E (2011) Alstroemeria hookeri Lodd. subsp. sansebastiana C.M. Baeza & E. Ruiz, nueva para la flora de Chile. Gayana Bot 68:313-315.
- Baeza C, Schrader O, Ruiz E and Negritto M (2007a) Análisis comparativo del cariotipo en poblaciones de Alstroemeria aurea R. Graham (Alstroemeriaceae) de Chile. Gayana Bot 64:33-39.
- Baeza C, Schrader O and Budahn H (2007b) Characterization of geographically isolated accessions in five Alstroemeria L. species using FISH of tandemly repeated DNA sequences and RAPD analysis. Plant Syst Evol 269: 1-14.
- Baeza C, Schrader O, Ruiz E and Negritto M (2008) Alstroemeria presliana Herb. (Alstroemeriaceae) in Chile from a cytogenetic perspective. Chilean J Agric Res 68:328-333.
- Baeza C, Ruiz E and Negritto M (2010) Comparative karyotypic analysis in the Alstroemeria hookeri Lodd. (Alstroemeriaceae) complex sensu Bayer (1987). Genet Mol Biol 33:119-124.
- Baeza C, Finot L and Ruiz E (2015) Comparative karyotype analysis of population in the Alstroemeria presliana Herbert (Alstroemeriaceae) complex in Chile. Genet Mol Biol 38:199-104.
- Baeza C, Finot V, Ruiz E, Carrasco P, Novoa P and Rosas M (2016a) Análisis citotaxonómico del complejo Alstroemeria diluta E. Bayer (Alstroemeriaceae) de Chile. Gayana Bot 73:221-227.
- Baeza C, Finot V, Ruiz E, Carrasco P, Novoa P, Stuessy T and González A (2016b) Comparative karyotypic analysis and cytotaxonomy in the Alstroemeria ligtu L. (Alstroemeriaceae) complex of Chile. Braz J Bot 39:305-313.
- Bayer E (1987) Die Gattung Alstroemeria in Chile. Mitt Bot Staatssamml Muenchen 24:1-362.
- Buitendijk J and Ramanna M (1996) Giemsa C-banded karyotypes of eigth species of Alstroemeria L. and some of their hybrids. Ann Bot 78:449-457.
- Buitendijk J, Boon E and Ramanna M (1997) Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Ann Bot 79:343-353.
- Buitendijk J, Peters A, Jan-Quené R and Ramanna M (1998) Genome size variation and C-band polymorphism in Alstroemeria aurea, A. ligtu and A. magnifica (Alstroemeriaceae). Plant Syst Evol 212:87-106.
- Cajas D, Baeza M, Ruiz E and Negritto M (2009) Análisis citogenético en poblaciones de Alstroemeria hookeri Lodd. subsp. hookeri (Alstroemeriaceae) en la Región del Bío-Bío, Chile. Gayana Bot 66:117-126.
- Chacón J, De Assis M, Meerow A and Renner S (2012a) From east Gondwana to Central America: Historical biogeography of the Alstroemeriaceae. J Biogeogr 39:1806-1818.
- Chacón J, Sousa A, Baeza C and Renner S (2012b) Ribosomal DNA distribution and a genus-wide phylogeny reveal patterns of chromosomal evolution in Alstroemeria (Alstroemeriaceae). Am J Bot 99:1501-1512.
- Eyzaguirre M (2008) Validación de Alstroemeria citrina Phil. (Alstroemeriaceae). Gayana Bot 65:241-244.
- Greilhuber J and Speta F (1976) C-banded karyotypes in the Scilla hohenackeri group S. persica, and Poschkina (Liliaceae). Plant Syst Evol 126:149-188.
- Hoffmann A, Watson J and Flores A (2015) Flora silvestre de Chile. Cuando el desierto florece. Volumen 1. Monocotiledóneas y otros taxones. Fundación Claudio Gay. Santiago, Salesianos impresores S.A. 261 p.
- Hofreiter A (2007) Biogeography and ecology of the Alstroemeriaceae–Luzuriagaceae clade in the high-mountain regions of Central and South America. Harv Pap Bot 12:259-284.
- Houben A, Wanner G, Hanson L, Verlin D, Leach C and Timmis J (2000) Cloning and characterization of polymorphic heterochromatic segments of Brachycome dichromosomatica Chromosoma 109:206-213.
- Levan A, Fredga K and Sandberg A (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201-220.
- Muñoz M (2003) Notas nomenclaturales sobre Alstroemeria L. (Alstroemeriaceae) Not Mens Mus Hist Nat 352: 22.
- Muñoz M and Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p.
- Muñoz M, Morales V and Moreira A (2011) Validación de Alstroemeria parvula Phil. (Alstroemeriaceae). Gayana Bot 68:114-116.
- Negritto M, Baeza C, Ruiz E and Novoa P (2015) Alstroemeria marticorenae (Alstroemeriaceae), a new species from central Chile. Syst Bot 40:69-74.
- Peruzzi L and Erôglu H (2013) Karyotype asymmetry: again, how to measure and what to measure? CompCytogen 7:1-9.
- R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
» https://www.R-project.org/ - Reeves A (2001) MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 44:239-443.
- Soetaert K (2016) plot3D: Plotting Multi-Dimensional Data. R package version 1.1. https://CRAN.R-project.org/package=plot3D
» https://CRAN.R-project.org/package=plot3D
-
Associate Editor: Marcelo Guerra
Publication Dates
-
Publication in this collection
14 May 2018 -
Date of issue
Apr./June 2018
History
-
Received
30 May 2017 -
Accepted
17 Nov 2017