Abstracts
This study reports the description of the karyotype of Mugil incilis from Venezuela. The chromosome complement is composed of 48 acrocentric chromosomes, which uniformly decrease in size. Therefore, the homologues can not be clearly identified, with the exception of one of the largest chromosome pairs, classified as number 1, whose homologues may show a subcentromeric secondary constriction, and of chromosome pair number 24, which is considerably smaller than the others. C-banding showed heterochromatic blocks at the centromeric/pericentromeric regions of all chromosomes, which were more conspicuous on chromosomes 1, given the C-positive signals include the secondary constrictions. AgNO3 and fluorescent in situ hybridization (FISH) with 45S rDNA demonstrated that the nucleolus organizer regions are indeed located on the secondary constrictions of chromosome pair number 1. FISH with 5S rDNA revealed that the minor ribosomal genes are located on this same chromosome pair, near the NORs, though signals are closer to the centromeres and of smaller size, compared to those of the major ribosomal gene clusters. This is the first description of co-localization of major and minor ribosomal genes in the family. Data are discussed from a cytotaxonomic and phylogenetic perspective.
C-banding; COI; 5S rDNA; Mullet; NORs
Se presenta la primera descripción del cariotipo de Mugil incilis de Venezuela. El complemento cromosómico está compuesto por 48 cromosomas acrocéntricos uniformemente decrecientes en tamaño. Por lo tanto, los homólogos no pueden ser claramente identificados, con excepción de uno de los pares de mayor tamaño, clasificado como número 1, cuyos homólogos poseen una constricción secundaria subcentromérica, y el par de cromosomas número 24, considerablemente más pequeño que los otros. El bandeo-C reveló bloques heterocromáticos en las regiones centroméricas/pericentroméricas de todos los cromosomas, más conspicuas en el cromosoma 1 en el que las señales C-positivas se encuentra localizada precisamente en la constricción secundaria. La tinción con AgNO3 y la Hibridación Fluorescente in situ (FISH) con sonda 45S rDNA revelaron que las regiones organizadoras del nucléolo están ciertamente localizadas sobre la constricción secundaria del cromosoma número 1. FISH con 5S rDNA reveló que los genes ribosomales menores están ubicados en este mismo par cromosómico, en posición proximal a las NORs, aunque cercanas al centrómero y de menor tamaño en comparación con los clúster de genes ribosomales mayores. Ésta es la primera descripción de co-localización de genes ribosomales mayores y menores en la familia Mugilidae. Los datos se discuten bajo perspectivas citotaxonómicas y filogenéticas.
Karyotype characterization of Mugil incilis Hancock, 1830 (Mugiliformes: Mugilidae), including a description of an unusual co-localization of major and minor ribosomal genes in the family
Anne Kathrin HettI; Mauro NirchioII; Claudio OliveiraIII; Zoila Raquel SicchaIII; Anna Rita RossiI; Luciana SolaI
IDepartment of Human and Animal Biology, University of Rome "La Sapienza". Via Borelli 50, 00161 Rome, Italy
IIEscuela de Ciencias Aplicadas del Mar, Universidad de Oriente, Isla de Margarita, Venezuela. nirchio@cantv.net
IIIDepartamento de Morfologia, Instituto de Biociências, Universidade Estadual Paulista. 18618-000 Botucatu, SP, Brazil
ABSTRACT
This study reports the description of the karyotype of Mugil incilis from Venezuela. The chromosome complement is composed of 48 acrocentric chromosomes, which uniformly decrease in size. Therefore, the homologues can not be clearly identified, with the exception of one of the largest chromosome pairs, classified as number 1, whose homologues may show a subcentromeric secondary constriction, and of chromosome pair number 24, which is considerably smaller than the others. C-banding showed heterochromatic blocks at the centromeric/pericentromeric regions of all chromosomes, which were more conspicuous on chromosomes 1, given the C-positive signals include the secondary constrictions. AgNO3 and fluorescent in situ hybridization (FISH) with 45S rDNA demonstrated that the nucleolus organizer regions are indeed located on the secondary constrictions of chromosome pair number 1. FISH with 5S rDNA revealed that the minor ribosomal genes are located on this same chromosome pair, near the NORs, though signals are closer to the centromeres and of smaller size, compared to those of the major ribosomal gene clusters. This is the first description of co-localization of major and minor ribosomal genes in the family. Data are discussed from a cytotaxonomic and phylogenetic perspective.
Key words: C-banding, COI, 5S rDNA, Mullet, NORs.
RESUMO
Se presenta la primera descripción del cariotipo de Mugil incilis de Venezuela. El complemento cromosómico está compuesto por 48 cromosomas acrocéntricos uniformemente decrecientes en tamaño. Por lo tanto, los homólogos no pueden ser claramente identificados, con excepción de uno de los pares de mayor tamaño, clasificado como número 1, cuyos homólogos poseen una constricción secundaria subcentromérica, y el par de cromosomas número 24, considerablemente más pequeño que los otros. El bandeo-C reveló bloques heterocromáticos en las regiones centroméricas/pericentroméricas de todos los cromosomas, más conspicuas en el cromosoma 1 en el que las señales C-positivas se encuentra localizada precisamente en la constricción secundaria. La tinción con AgNO3 y la Hibridación Fluorescente in situ (FISH) con sonda 45S rDNA revelaron que las regiones organizadoras del nucléolo están ciertamente localizadas sobre la constricción secundaria del cromosoma número 1. FISH con 5S rDNA reveló que los genes ribosomales menores están ubicados en este mismo par cromosómico, en posición proximal a las NORs, aunque cercanas al centrómero y de menor tamaño en comparación con los clúster de genes ribosomales mayores. Ésta es la primera descripción de co-localización de genes ribosomales mayores y menores en la familia Mugilidae. Los datos se discuten bajo perspectivas citotaxonómicas y filogenéticas.
Introduction
The fish family Mugilidae (mullets) is composed of approximately 70 species and it inhabits coastal and brackish waters of all tropical and temperate regions of the world (Thomson, 1997; Froese & Pauly, 2010). Recent cytogenetic studies carried out on several Mugilidae species from Venezuela (reviewed in Sola et al., 2007, 2008) have provided information regarding several interesting systematic and cytotaxonomic issues within the family. For example, the cytogenetic studies made it possible to establish that Mugil rubrioculus is a separate species from M. curema (Nirchio et al., 2003, 2007) and also indicated the likely existence of a species complex within M. curema (Nirchio & Cequea, 1998; Nirchio et al., 2005a). Moreover, the localization of major and minor ribosomal genes revealed that the 48 uniarmed chromosome karyotype, displayed by all the Mugilidae species, with the exception of M. curema, is much more differentiated than the macromorphology of chromosomes would suggest (Sola et al., 2007, 2008).
According to Cervigón (1993), in Venezuela the family Mugilidae is represented by six mullet species, four of which, Mugil curema, M. rubrioculus (identified as M. gaimardianus), M. liza, M. trichodon, have already been cytogenetically investigated (reviewed in Sola et al., 2007, 2008). The karyotype of M. curvidens has yet to be described, while for M. incilis little chromosome information is available for specimens from Brazil (Pauls et al., 1998; Galetti et al., 2000; Netto et al., 2007).
This study was carried out to continue the karyological characterization of Venezuelan mugilids, presenting cytogenetic data of the parassi mullet, Mugil incilis. This species lives in brackish estuaries but also in the marine and hyper-saline waters (Cervigón et al., 1992) of the Western Atlantic from the West Indies and the Atlantic coasts of Central America southward to Brazil (Froese & Pauly, 2010). This paper reports the chromosome number and formula, constitutive heterochromatin distribution by C-banding and the locations of major and minor ribosomal genes by Ag-staining and Fluorescent in situ Hybridization (FISH) in M. incilis.
Material and Methods
A total of nine specimens of Mugil incilis were caught with seine nets in La Restinga Lagoon, Venezuela, in March 2009. Morphological species identification was based on Cervigón (1993) and Harrison (2002). For a subset of five specimens, a fragment of cytochrome c oxidase subunit I (COI) mitochondrial gene was amplified and sequenced in order to evaluate the species identification. For this molecular analysis white muscle tissue was excised from fishes and preserved in 95% alcohol. DNA isolation and polymerase chain reaction followed the procedures reported in Ward et al. (2005). Voucher specimens were deposited at the Ichthyology Collection of the Escuela de Ciencias Aplicadas del Mar, Universidad de Oriente (ECAM 780-788).
Chromosome preparations were obtained from cephalic kidney cells using conventional air-drying techniques (Foresti et al., 1993). Silver-stained nucleolus organizer regions (Ag-NORs) were obtained by chromosome impregnation with silver nitrate as described by Howell & Black (1980). C-bands were obtained following the method of Sumner (1972).
The 45S and 5S rDNA clusters were mapped by FISH following the method of Pinkel et al. (1986). The 45S rDNA probe (kindly provided by Angelo Libertini) contained the 18S-5.8S-28S genes plus an intergenic spacer from Drosophila melanogaster and was labelled by random priming with biotin using the BioPrime Labeling system (Invitrogen). The 5S rDNA repeat probe was generated by PCR with the primers 5SA (5'TAC GCC CGA TCT CGT CCG ATC3') and 5SB (5'CAG GCT GGT ATG GCC GTA AGC3') (Martins & Galetti Jr., 1999) from genomic DNA of Squalius lucumonis and directly labelled with biotin-14-dATP. Post-hybridization washes were carried out at low stringency (37ºC, 10 min each). Signals were detected and amplified by a two-round application of Avidin-FITC/biotinilated Anti-avidin (Vector). Metaphases were counterstained with 4',6-diamidino-2-phenylindole (DAPI) and propidium iodide, and examined under a Zeiss Axiophot epifluorescence microscope equipped with the appropriate selective filters. The images were acquired using a SenSys 1400 CCD camera and then artificially coloured and elaborated with Photoshop CS3 (Adobe Systems Inc).
Results
The COI fragments obtained from PCR amplification from five specimens ranged from 567 to 654 nucleotides in size. Sequences were deposited in GenBank (accession number HQ285927- HQ285931) and blasted in the Barcode of Life Data Systems (BOLD). Sequence similarity (calculated on the common 567 nucleotides) was 99.8% among the five M. incilis specimens analyzed and up to a maximum of 94.5% with M. curema and M. liza (identified as M. platanus) and of 94.66% with M. trichodon. In all analysis, the M. incilis specimens form a separate lineage from the other Mugil species with sequences already included in the Genbank and BOLD.
In all the nine parassi mullet specimens examined, the karyotype (Fig. 1) was found to be made of 2n=48 acrocentric chromosomes (Fundamental Number of arms, FN=48). Chromosomes uniformly decrease in size, making it difficult to clearly identify the homologues, with the exception of one of the largest chromosome pairs, classified as number 1, whose homologues may show a more or less pronounced subcentromeric secondary constriction, and of chromosome pair number 24, which is considerably smaller than the others.
C-banding (Fig. 2a), sequential to Giemsa-staining (not shown), revealed a prominent C-positive block on chromosome pair number 1, which seems to include both the centromeric/pericentromeric heterochromatin and the secondary constrictions themselves. All remaining chromosomes show C-positive heterochromatin in the centromeric/pericentromeric regions. Ag-staining revealed the Ag-NORs (Fig. 2b), located on the secondary constriction of chromosome pair number 1.
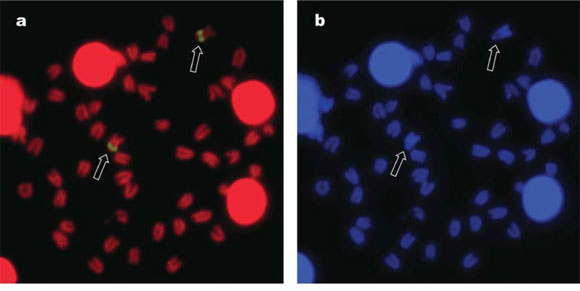
FISH with a 45S rDNA probe produced bright signals on the same subcentromeric location of the Ag-positive sites (Fig. 3a), and no additional NOR-sites could be identified. On the same DAPI- counterstained metaphase plate (Fig. 3b), the NOR-bearing chromosome pair can be easily identified by the DAPI-negative region in subcentromeric position, corresponding to the secondary constriction. FISH with a 5S rDNA probe (Fig. 3c) produced hybridization signals on this same chromosome pair (Fig. 3d), again in a subcentromeric location. However, the signals are closer to the centromeres and smaller in size compared to those of the major ribosomal gene clusters.
Discussion
In order to allow future comparison of the cytogenetic features here reported for the Venezuela specimens of M. incilis with specimens from different localities, five specimens were sequenced and the sequences of COI region were deposited in the GenBank. After blasting in Genbank and BOLD, both the level of sequence similarity and the tree topology reveal that the specimens analyzed in present study belong to a separate species from all the Mugil species already investigated through this important molecular marker (M. curema, M. liza, and M. trichodon), which is a useful complement to their morphological identification as M. incilis.
Eighteen species of the family Mugilidae have so far been cytogenetically analysed and seven of these belong to genus Mugil. As previously described (Sola et al., 2007, 2008), four main cytotypes (A, B, C1, and C2) can be distinguished in mullet species. The chromosome complement of M. incilis reported in this study conforms to the cytotype A, with exclusively acrocentric chromosomes, which is shared by almost all the investigated Mugil species. Noticeably, M. curema shows a reduced diploid number but a conserved FN (=48) and, as a consequence, two cytotypes, mainly (cytotype C1, 2n=28, in specimens from Brazil and from the Gulf of Mexico) or exclusively (cytotype C2, 2n=24, in specimens from Venezuela) composed of bi-armed chromosomes. The remaining 11 Mugilidae species also show the conservative 48 uniarmed karyotype. However, while five of them show cytotype A, the six remaining species show cytotype B, which differs from the former due to the presence of a single subtelocentric chromosome pair among the remaining acrocentrics of the chromosome complement.
Present results disagree with the karyological data previously reported for the same species. Indeed, in a review on marine fish cytogenetic (Galetti et al., 2000), a conference communication was quoted (Pauls et al., 1998) reporting a significant reduction in the number of chromosomes (2n=28) in specimen(s) of M. incilis from Brazil. Lately, a metaphase plate, mainly composed of bi-armed chromosomes, and with the same chromosome number (2n=28), was shown by the same research group (Netto et al., 2007) for one M. incilis individual, in a technical paper on chromosome preparation. Unfortunately, no precise information is available neither on sampling localities (along the Atlantic coast of Bahia and/or Rio de Janeiro States), nor on the specimens classification criteria. Moreover the number of examined specimens is also unknown for Pauls et al. (1998).
Thus, at present it is not possible to speculate whether the karyotype differences among Venezuelan and Brazilian specimens reflect an actual intraspecific polymorphism, or the existence of cryptic species, or are instead due to a misclassification of Brazilian samples. As a matter of fact, the conservative external morphology of the family is at the basis of many systematic problems, as evidenced by the reduction of 233 nominal species to only 80 valid species (Pauly & Froese, 2010). Specifically considering mullet in the area, through different mtDNA markers (Fraga et al., 2007; Heras et al., 2009; BOLD), at least three different clusters can be identified within M. curema, which also include other mullet species. Similarly, in the same species complex, from a cytogenetic point of view, in addition to the two cytotypes described above, it has to be mentioned the chromosome complement of M. rubrioculus, formerly considered M. curema (Nirchio et al., 2003, 2007), which has allowed to confirm it as a valid species. As one M. curema population from Brazil (Nirchio et al., 2005) exhibits a chromosome complement with 2n=28 (Cytotype 1) apparently similar to the one described for M. incilis from Brazil (Pauls et al., 1998; Galetti et al., 2000; Netto et al., 2007), further sampling of Brazilian specimens is necessary to check out whether or not they belong to a different species from M. curema.
The analysis of the constitutive heterochromatin distribution revealed a generalized centromeric/pericentromeric location. This analysis has been so far applied to a limited number (ten out of 17) of the Mugilidae species with the 48 uniarmed karyotype, but the location appear to be the same in all of them. Nevertheless, in M. incilis the conspicuous C-positive block in chromosome pair number 1 doubtlessly identifies this pair as a peculiar species-specific chromosome marker. Indeed the larger amount of C-positive heterochromatin in this chromosome pair was found to be associated with the syntenic and contiguous location of both minor and major ribosomal gene clusters in this chromosome region.
The NOR sites, so far investigated in 12 out of the 17 mullet species with the 48 uniarmed karyotype, have been mostly localized either in terminal chromosomal regions or on the short arms of the single subtelocentric chromosome pair of the six species with cytotype B (Sola et al., 2007, 2008). Only in two species, M. trichodon (Nirchio et al., 2005b) and M. rubrioculus (Nirchio et al., 2007), the NOR sites show an interstitial location. However, in these latter species, the NOR-bearing chromosomes are of medium size and smaller than the NOR-bearing chromosome pair of M. incilis. Thus, this NOR-location represents a newly detected feature in the family.
As far as the 5S rDNA sites are concerned (investigated in eight out of the 17 species with the 48 uni-armed karyotype), they have been localized in an interstitial position of a medium-sized chromosome pair in most (seven) of the species. In Liza saliens, in addition to this, a second site for 5S rDNA was detected, located in the terminal position of chromosome pair number 1 (Gornung et al., 2001). Therefore, also the 5S rDNA location observed in M. incilis is new for the family.
However, what is unique is that in M. incilis, differently from all Mugilidae species investigated in this sense, the two classes of ribosomal genes are co-localized on the same chromosome pair. In fact, the major and 5S rDNA seem to occupy contiguous regions of the homologues 1, the former apparently confined to the achromatic secondary constriction, the latter flanking the clusters of 45 S rDNA and located in the euchromatic region, closer to the centromere.
The chromosomal location of the 5S rRNA genes has been investigated in over 60 fish species (Martins, 2007), and the location of minor and major rDNA clusters on different chromosome pairs is the more common condition found in bony fish, including those species which have more than one chromosomal site for 5S rDNA. However, evidence of co-localization of 5S and 18S rDNA has been reported for some species of Acipenseriformes (Fontana et al., 2003), Salmoniformes (e.g., Pendas et al., 1994; Moran et al., 1996; Fujiwara et al., 1998; Rossi & Gornung, 2005), Cypriniformes (Inafuku et al., 2000), Characiformes (e.g., Almeida-Toledo et al., 2002; Cioffi et al., 2009; Diniz et al., 2009 and references therein), Perciformes (Nirchio et al., 2009; Pisano & Ghigliotti, 2009) and, now also, Mugiliformes (present study). Interestingly, all the possible syntenic conditions have been found, either the two sets of genes in distinct and disjoint chromosomal regions, as in Astyanax scabripinnis (Mantovani et al., 2005) and Parodon tortuosus (=Parodon nasus) (Vicente et al., 2001), or in adjacent regions, as in M. incilis (present study) and Triportheus nematurus (Diniz et al., 2009), or the 5S rRNA genes interspersed along the clusters of 45S rDNA, as in Notothenioidei (Pisano & Ghigliotti, 2009).
The peculiar cytotaxonomic features of Mugil incilis might constitute a clear cut diagnostic tool in showing species boundaries, which might be very helpful in identifying cryptic species in the M. curema complex, as already occurred (Nirchio et al., 2003, 2007); on the other hand, present data indicates that it would be useful to add cytogenetic data on the other Mugilidae species which have yet to be described and to parallely increase the molecular database on them, in order to clarify the karyoevolutive patterns and the phylogenetic relationships within the family.
Acknowledgements
Financial support was provided by Consejo de Investigación, Universidad de Oriente, Venezuela, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil and "La Sapienza" University of Rome, Italy.
Literature Cited
Accepted November 17, 2010
- Almeida-Toledo, L., C. Ozouf-Costaz, F. Foresti, C. Bonillo, F. Porto-Foresti & M. F. Z. Daniel-Silva. 2002. Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae). Cytogenetic and Genome Research, 97: 229-233.
- Cervigón, F. 1993. Los Peces Marinos de Venezuela. Volumen II. Caracas, Fundación Científica Los Roques, 499p.
- Cervigón, F., R. Cipriani, W. Fischer, L. Garibaldi, M. Hendrickx, A. J. Lemus, R. Márquez, J. M. Poutiers, G. Robaina & B. Rodriguez. 1992. Fichas FAO de identificación de especies para los fines de la pesca. Guía de campo de las especies comerciales marinas y de aquas salobres de la costa septentrional de Sur América. FAO, Rome, 513p.
- Cioffi, M. B., C. Martins, L. Centofante, U. Jacobina & L. A. C. Bertollo. 2009. Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: mapping of three classes of repetitive DNAs. Cytogenetic and Genome Research, 125: 132-141.
- Diniz, D., A. Laudicina & L. A. C. Bertollo. 2009. Chromosomal location of 18S and 5S rDNA sites in Triportheus fish species (Characiformes, Characidae). Genetics and Molecular Biology, 32: 37-41.
- Fontana, F., M. Lanfredi, L. Congiu, M. Leis, M. Chicca & R. Rossi. 2003. Chromosomal mapping of 18 S-28 S and 5 S rRNA genes by two-colour fluorescent in situ hybridization in six sturgeon species. Genome, 46: 473-477.
- Foresti, F., C. Oliveira & L. F. Almeida-Toledo. 1993 A method for chromosome preparations from large specimens of fishes using in vitro short treatment with colchicine. Experientia, 49: 810-813.
- Fraga, E., H. Schneider, M. Nirchio, E. Santa-Brigida, L. F. Rodrigues-Filho & I. Sampaio. 2007. Molecular phylogenetic analyses of mullets (Mugilidae, Mugiliformes) based on two mitochondrial genes. Journal of Applied Ichthyology, 23: 598-604.
- Froese, R. & D. Pauly. 2010. FishBase. World Wide Web electronic publication. Available at: http//www.fishbase.org/ Accessed October, 2010.
- Fujiwara, A., S. Abe, E. Yamaha, F. Yamazaki & M. C. Yoshida. 1998. Chromosomal localization and heterochromatin association of ribosomal RNA gene loci and silver-stained nucleolar organizer regions in salmonid fishes. Chromosome Research, 6: 463-471.
- Galetti, P. M. Jr., C. T. Aguilar & W. F. Molina. 2000. An overview of marine fish cytogenetics. Hydrobiologia, 420: 55-62.
- Gornung, E., C. A. Cordisco, A. R. Rossi, S. De Innocentiis, D. Crosetti & L. Sola. 2001. Chromosomal evolution in Mugilidae: karyotype characterization of Liza saliens and comparative localization of major and minor ribosomal genes in the six Mediterranean mullet. Marine Biology, 139: 55-60.
- Harrison, I. J. 2002. Mugilidae. Pp. 1071-1085. In: Carpenter, K. E. (Ed.). The Living Marine Resources of the Western Central Atlantic, Vol. 2. Bony Fishes part 1 (Acipenseridae to Grammatidae). FAO Species Identification Guide for Fisheries Purposes and American Society of Ichthyologists and Herpetologists Special Publication No. 5, pp. 601-1374. Rome, FAO.
- Heras, S., M. I. Roldan & M. G. Castro. 2009. Molecular phylogeny of Mugilidae fishes revised. Reviews in Fish Biology and Fisheries, 19: 217-231.
- Howell, W. M. & D. A. Black. 1980. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia, 36: 1014-1015.
- Inafuku, J., M. Nabeyama, Y. Kikuma, J. Saitoh, S. Kubota & S. Kohno. 2000. Chromosomal location and nucleotide sequences of 5S ribosomal DNA of two cyprinid species (Osteichthyes, Pisces). Chromosome Research, 8: 193-199.
- Mantovani, M., L. Douglas dos Santos, A. Moreira-Filho & O. Moreira-Filho. 2005. Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica, 123: 211-216.
- Martins, C. 2007. Chromosomes and repetitive DNAs: a contribution to the knowledge of the fish genome. Pp. 421-454. In: Pisano, E., C. Ozouf-Costaz, F. Foresti & B. G. Kapoor (Eds.). Fish Cytogenetics. Enfield, Science Publishers, 502p.
- Martins, C. & P. M. Galetti Jr. 1999. Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Research, 7: 363-367.
- Moran, P., J. Martinez, E. Garcia-Vazquez & A. Pendas. 1996. Sex linkage of 5 S rDNA in rainbow trout (Oncorhynchus mykiss). Cytogenetics and Cell Genetics, 75: 145-150.
- Netto, M. R. C. B., E. Pauls & P. R. A. M. Affonso. 2007. A standard protocol for obtaining fish chromosomes under post-mortem conditions. Micron, 38: 214-217.
- Nirchio, M. & H. Cequea. 1998. Karyology of Mugil liza and M. curema from Venezuela. Boletin de Investigaciones Marinas y Costeras, 27: 45-50.
- Nirchio, M., F. Cervigón, J. Porto, J. Pérez, J. A. Gómez & J. Villalaz. 2003. Cytogenetic confirmation of Mugil curema Valenciennes, 1836 and Mugil gaimardianus Desmarest, 1831 (Mugilidae: Teleostei) as valid nominal species. Scientia Marina, 67: 113-115.
- Nirchio, M., R. R. Cipriano, M. M. Cestari, & A. S. Fenocchio. 2005a. Cytogenetical and morphological features reveal significant differences among Venezuelan and Brazilian samples of Mugil curema Neotropical Ichthyology, 3: 107-110.
- Nirchio, M., C. Oliveira, I. A. Ferreira, J. E. Pérez, J. I. Gaviria, I. Harrison, A. R. Rossi & L. Sola. 2007. Comparative cytogenetic and allozyme analysis of Mugil rubrioculus and Mugil curema (Teleostei: Mugilidae) from Venezuela. Interciencia, 32: 757-762.
- Nirchio, M., C. Oliveira, D. C. Ferreira, R. Rondón, J. E. Pérez, A. K. Hett, A. R. Rossi & L. Sola. 2009. Cytogenetic characterization of Rhomboplites aurorubens and Ocyurus chrysurus, two monotypic genera of Lutjaninae from Cubagua Island, Venezuela, with a review of the cytogenetics of Lutjanidae (Teleostei: Perciformes). Neotropical Ichthyology, 7: 587-594.
- Nirchio, M., E. Ron & A. R. Rossi. 2005b. Karyological characterization of Mugil trichodon Poey, 1876 (Pisces: Mugilidae). Scientia Marina, 69: 525-530.
- Pauls, E., I. A. Coutinho, P. R. A. M. Affonso, M. R. C. B. Netto, A. S. S. Oliveira, W. Guedes & A. Afonso. 1998. Characterization of fish diversity along the coast of the state of Rio de Janeiro: a cytogenetic approach. First International Workshop on Marine Genetics - Rio 98, Rio de Janeiro, Brazil, book of abstracts: 58p.
- Pendas, A. M., P. Moran, J. P. Freije & E. Garcia-Vazquez. 1994. Chromosome mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenetics and Cell Genetics, 63: 31-36.
- Pinkel, D., T. Straume & J. W. Gray. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences, 83: 2934-2938.
- Pisano, E. & L. Ghigliotti. 2009. Ribosomal genes in notothenioid fishes: Focus on the chromosomal organisation. Marine Genomics, 2: 75-80.
- Rossi, A. R. & E. Gornung. 2005. Cytogenetic analysis of three Italian populations of Coregonus lavaretus (Pisces, Salmoniformes) with chromosomal location of major and minor ribosomal genes, and telomeric repeats. Hereditas, 142: 15-21.
- Sola, L., E. Gornung, M. E. Mannarelli & A. R. Rossi. 2007. Chromosomal evolution in Mugilidae, Mugilomorpha: an overview. Pp. 165-194. In: Pisano, E., C. Ozouf-Costaz, F. Foresti & B. G. Kapoor (Eds.). Fish Cytogenetics. Enfield, Science Publishers, 502p.
- Sola, L., M. Nirchio & A. R. Rossi. 2008. Past and future of cytogenetics of mugilidae: an updated overview. Boletin del Instituto Oceanográfico de Venezuela, 47: 25-31.
- Sumner, A. T. 1972. A simple technique for demonstrating centromeric heterocromatin. Experimental Cell Research, 75: 304-306.
- Thomson, J. M. 1997. The Mugilidae of the world. Memoirs of the Queensland Museum, 41: 457-562.
- Vicente, V. E., C. M. Jesus & O. Moreira-Filho. 2001. Chromosomal localization of 5S and 18S rRNA genes in three Parodon species (Pisces, Parodontidae). Caryologia, 54: 365-369.
- Ward, R. D., T. S. Zemlak, B. H. Innes, P. R. Last & P. D. N. Hebert. 2005. DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society, Series B, 359: 1847-1857.
Publication Dates
-
Publication in this collection
04 Feb 2011 -
Date of issue
Mar 2011