Abstracts
The objective of this study was to establish a specific, sensitive and rapid PCR approach for the detection of Clostridium sp. at the genus level. Clostridium sp. in the duodenum, jejunum, ileum and cecum of broiler chickens were analyzed by 16S rRNA genes. The PCR detected the presence of Clostridium spp. in naturally contaminated intestinal samples. For the total gastrointestinal segments, 53.125, 65.625 and 59.375% samples were positive for naturally occurring Clostridium spp. at the ages 4, 14 and 30d, respectively. Analysis of the microbial contents indicated that Clostridium sp. was not consistently detected in all intestinal segments. These results can put in evidence the hypothesis that Clostridium spp. may be interfering in health and performance of chickens.
Clostridium spp.; broiler; gastrointestinal; age; 16S rRNA genes
Clostridium spp. são organismos patogénicos com distribuição mundial, podendo estar presente nos seres humanos, em animais domésticos e em animais selvagens. Estas bactérias habitam geralmente o trato gastrintestinal. Os métodos bacteriológicos convencionais como a microscopia e a cultura têm limitações. O objetivo deste estudo foi de estabelecer uma metodologia específica, sensível e rápida como a ténica de PCR para a deteção de Clostridium spp. A presença de Clostridium spp. Foi pesquisada no duodeno, o jejunum, o íleo e o cecum de galinhas usando análise molecular de genes do rRNA 16S. A técnica de PCR usada neste trabalho detectou Clostridium spp. em amostras intestinais naturalmente contaminadas. Considerando o trato gastrintestinal total, 53.125, 65.625 e 59.375% das amostras foram positivas para Clostridium nas idades 4, 14 e 30d respectivamente. A análise microbiana indicou que Clostridium spp. não foi detectado consistentemente em todos os segmentos intestinais. Os dados observados alertam para possíveis implicações significativas para a saúde e o desempenho das galinhas.
HUMAN AND ANIMAL HEALTH
Detection of Clostridium sp. and its Relation to Different Ages and Gastrointestinal Segments as Measured by Molecular Analysis of 16S rRNA Genes
Seyed Ziaeddin MirhosseiniI, II; Alireza SeidaviIII, * * Author for correspondence: alirezaseidavi@yahoo.com ; Mahmoud ShivazadIV; Mohammad ChamaniV; Ali Asghar SadeghiV; Reza PourseifyII
IAnimal Science Department; Guilan University; Rasht - Iran
IIAgriculture Biotechnology Research Institute of North Region of IRAN; Iran
IIIAnimal Science Department; Islamic Azad University; Rasht Branch; Rasht - Iran
IVAnimal Science Department; Tehran University; Karaj; Iran
VAnimal Science Department; Islamic Azad University; Science and Research Branch; Tehran - Iran
ABSTRACT
The objective of this study was to establish a specific, sensitive and rapid PCR approach for the detection of Clostridium sp. at the genus level. Clostridium sp. in the duodenum, jejunum, ileum and cecum of broiler chickens were analyzed by 16S rRNA genes. The PCR detected the presence of Clostridium spp. in naturally contaminated intestinal samples. For the total gastrointestinal segments, 53.125, 65.625 and 59.375% samples were positive for naturally occurring Clostridium spp. at the ages 4, 14 and 30d, respectively. Analysis of the microbial contents indicated that Clostridium sp. was not consistently detected in all intestinal segments. These results can put in evidence the hypothesis that Clostridium spp. may be interfering in health and performance of chickens
Key words:Clostridium spp., broiler, gastrointestinal, age, 16S rRNA genes
RESUMO
Clostridium spp. são organismos patogénicos com distribuição mundial, podendo estar presente nos seres humanos, em animais domésticos e em animais selvagens. Estas bactérias habitam geralmente o trato gastrintestinal. Os métodos bacteriológicos convencionais como a microscopia e a cultura têm limitações. O objetivo deste estudo foi de estabelecer uma metodologia específica, sensível e rápida como a ténica de PCR para a deteção de Clostridium spp. A presença de Clostridium spp. Foi pesquisada no duodeno, o jejunum, o íleo e o cecum de galinhas usando análise molecular de genes do rRNA 16S. A técnica de PCR usada neste trabalho detectou Clostridium spp. em amostras intestinais naturalmente contaminadas. Considerando o trato gastrintestinal total, 53.125, 65.625 e 59.375% das amostras foram positivas para Clostridium nas idades 4, 14 e 30d respectivamente. A análise microbiana indicou que Clostridium spp. não foi detectado consistentemente em todos os segmentos intestinais. Os dados observados alertam para possíveis implicações significativas para a saúde e o desempenho das galinhas.
INTRODUCTION
Poultry farming is a worldwide practice of meat production that has significantly increased in the last few decades. Microbial flora in chicken gastrointestinal is important to monitor both for animal welfare and food safety reasons (Apajalahti et al. 1998). The microflora plays an important role through their effects on gut morphology, nutrition, immune responses and protection against colonisation of pathogens (Hill et al., 2002; Lu et al. 2003).
In the poultry industry, some Clostridium spp. e.g. Clostridium perfringens are important and notable species of Clostridium genus. Clostridium type A or C in poultry are important pathogens that colonize the gastrointestinal (GI) tract of chickensis and are the causative agent of necrotic enteritis and sub-clinical disease (Ficken and Wages, 1997; Tech, 1999; Shanmugavelu et al., 2006; Barbara et al., 2007). Both C perfringens types A and C are associated with necrotic enteritis in poultry, a disease of considerable economic significance to the poultry industry due to increased mortality and reduced weight gain (La-Ragione and Woodward, 2003; McCourt et al., 2005). The alpha-toxin producing C perfringens is a phospholipase C sphingomyelinase that hydrolyzes phospholipids, and as a consequence induces the production of inflammatory mediators (Titball, 1993). This potentially leads the blood vessel contraction and myocardial dysfunction, and as a consequence acute death (Olkowski et al. 2006). Therefore, C perfringens, using this toxin, causes necrotic enteritis (NE) in chickens, which can lead to increased mortality, impaired feed conversion, and retarded growth rate (Lovland and Kaldhusdal, 1999; Petit et al. 1999; Kaldhusdal et al. 2001). There is mounting evidence that with the withdrawal of in-feed antibiotics, the incidence of this disease will increase (Grave et al., 2004).
While the estimate of cultivability of GI bacteria e.g. Clostridium spp. is relatively high compared to most microbial ecosystems, the culturable fraction is still a minority. The reasons for this cultivation anomaly include the unknown growth requirements of the bacteria, the selectivity of the media that are used, the stress imposed by the cultivation procedures, the necessity of strictly anoxic conditions, and difficulties with simulating the interactions of bacteria with other microbes and host cells. The circumvention of these limitations requires culture-independent methods (Zoetendal et al., 2004). A dramatic increase in the application of approaches based on the sequence diversity of the 16S ribosomal RNA (rRNA) gene have been made during the past decade to explore the diversity of bacterial communities in a variety of ecosystems, including the mammalian GI tract. Analyses of bacterial rDNA sequences from chicken fecal DNA extracts suggest that the chicken cecum and ileum are inhabited by a diverse bacterial community (Zhu et al., 2002; Lu et al., 2003; Gholamiandekhordi et al., 2006). Today, the PCR has become a powerful and increasingly popular tool in microbial identification (Nauerby et al. 2003; Skansenga et al. 2006; Lu et al. 2007). The great advantage of the PCR procedure is that it can be applied to mixed microbial specimens without prior isolation of individual species of bacteria. The potential of PCR to detect genetic sequences from minute quantities of DNA is advantageous compared to microbiologic and serologic detection methods. PCR assays to detect genera or species in gastrointestinal or other samples have sensitivity and specificity despite the high density and diversity of the native microflora in the samples. Molecular detection of the Clostridium spp. by polymerase chain reaction (PCR) is one useful method for detecting Clostridium spp. in some samples such as food raw materials and human and animal fecal (Engström et al. 2003; Carlin et al. 2004; Jaimes et al. 2006).
In view of the above, the detection of Clostridium sp. using PCR-based method for gastrointestinal contents of broiler at three different ages of rearing period was investigated.
MATERIALS AND METHODS
Broiler maintenance and sample collection
Thirty six Commercial broilers (Ross 308) were raised under conditions identical to those found in commercial broiler operations. The broilers were not exposed to competitive exclusion preparations as newly hatched chicks and were fed a diet of commercial feed (NRC, 1994). At the ages 4, 14 and 30d, eight birds were randomly selected and sacrificed by cervical dislocation. Then duodenum, jejunum, ileum and cecum were removed aseptically, clamped with forceps, and placed in sterile plastic bags on ice. In the laboratory, each section was inverted onto sterile glass rods. Approximately one gram of content were collected into a centrifuge tube containing 9 ml of sterile phosphate-buffered saline (PBS, pH 7.4), and homogenized by vortexing with glass beads (4-mm diameter) for 3 min. Debris was removed by centrifugation at 700g for 1 min, and the supernatant were centrifuged at 13,000g for 5 min. The pellet was washed twice with PBS and stored at -20°C until DNA extraction.
Two type samples were used in the PCR amplification including 96 individual samples (included 4 gastrointestinal segments × 3 different ages × 8 replications or chicks) and 12 mixed samples (included 4 gastrointestinal segments × 3 different ages). In 12 mixed samples, all eight similar replications or chicks' samples were mixed together.
DNA extraction and preparation
Bacterial genomic DNA was isolated by the method of Seidavi et al (2007) with some modifications. Briefly, samples were centrifuged at 14,500g for 2 min and the cells were re-suspended thoroughly in 480μl of 50mM EDTA. Then 60μl of 10mg/ml Lysosyme enzyme was added. The samples were incubated at 37°C for 45 min and centrifuged at 14,500g for 2 min. Then after adding 600μl Nuclei Lysis Solution was and incubation at 80°C for 5 min, 3μl of RNase Solution was added and samples incubated at 37°C for 30 min. After that, 200μl of Protein Precipitation Solution was added to RNase-treated cell lysate. Samples were incubated on ice for 5 min and centrifuged at 14,500g for 3 min and the supernatant was transferred to 1.5ml microcentrifuge tubes containing 600μl of isopropanol. Tubes were centrifuged at 14,500g for 2 min. The supernatant were poured off and 600μl of 70% ethanol was added. Tubes were centrifuged at 14.500g for 2 min and ethanol was carefully aspirated. Then 100μl of DNA Rehydration Solution were added to the tubes and the DNA was rehydrated by incubating at 65°C for 60 min. Thus prepared DNA was stored at 4°C till PCR amplification.
PCR amplification
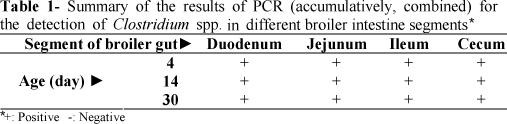
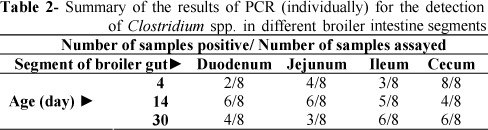
The PCR amplification mixtures (25 μl) consisted of 1μl of 25 ng DNA sample, 0.08 mM of each dNTP, 1.2 mM MgCl2, 1× PCR buffer, 0.28 ρM of each primer (forward and reverse), 1 U of Taq DNA polymerase and 18.6μl ddH2O. Amplification was performed on a thermocycler (ABI 9700) where initial denaturation was at 95°C for 3 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 54°C for 40 s and extension at 72°C for 80 s, with a final extension at 72°C for 3 min. Amplified products were electrophoresed in 2% agarose gels containing ethidium bromide. A pUC Mix Marker 8 was used as molecular size marker. Experiment was conducted both individual DNA samples (Table 2 and Fig 2) and also DNA of eight same samples which combined together (Table 1 and Fig 1).
PCR assay
The specific detection of the Clostridium sp. was based on PCR amplification of the 16S rRNA gene using oligonucleotide primers Clos58-f AAAGGAAGATTAATACCGCATAA and Clos780-r ATCTTGCGACCGTACTCCCC which PCR product size was 722 bp as described by Amit-Romach et al. (2004). The GenBank program BLAST was used to ensure that the applied primers were complementary with the target species but not with other species. Primers were compared with sequences in the GenBank, and none were found to have the exact same sequence as the non-targeted sequence.
Positive and negative Controls
In both individual and mixed sample procedures, negative and positive controls were used for Clostridium sp. confirmation and detection by PCR technique. ddH2O was used as negative control to confirm the absence of contamination of material and facilities and removal of experimental errors and to prove the exclusion of non-target DNA. The C perfringens strain as positive control used in this study was prepared from the bacterial isolate archive of the Razi Institute of Iran. Double-stranded DNA extracted from each isolate was examined along PCR to confirm the presence of PCR-compatible DNA.
RESULTS
A total of 96 bird/gut segment samples including 24 duodenum (4, 14 and 30d), 24 jejunum (4, 14 and 30d), 24 ileum (4, 14 and 30d) and 24 cecum (4, 14 and 30d) were examined by the molecular method for Clostridium sp. detection (see Tables 1 and 2 and Figs 1 and 2). For total intestine segments, a total of 57 samples were positive for naturally occurring Lactobacillus sp. by the PCR method. This value represented an incidence of 59.375% with respect to the total gut samples. Furthermore, for total intestine segments, a total of 17, 21 and 19 samples were positive for naturally occurring Clostridium spp. by PCR method at 4, 14 and 30d respectively. These values represented an incidence of 53.125, 65.625 and 59.375% with respect to the total gut samples, respectively.
For duodenum, a total of 2, 6 and 4 samples were positive for naturally occurring Clostridium sp. by PCR method at 4, 14 and 30d, respectively. These values represented an incidence of 25, 75 and 50% with respect to the total gut samples, respectively.
For jejunum, a total of 4, 6 and 3 samples were positive for naturally occurring Clostridium sp. by PCR method at 4, 14 and 30d, respectively. These values represented an incidence of 50, 75 and 37.5% with respect to the total gut samples, respectively.
For ileum, a total of 3, 5 and 6 samples were positive for naturally occurring Clostridium sp. by PCR method at 4, 14 and 30d, respectively. These values represented an incidence of 37.5, 62.5 and 75% with respect to the total gut samples, respectively.
For cecum, a total of 8, 4 and 6 samples were positive for naturally occurring Clostridium sp. by PCR method at 4, 14 and 30d, respectively. These values represented an incidence of 100, 50 and 75% with respect to the total gastrointestinal samples, respectively. These percentages are in agreement with the frequency of Clostridium sp. among the intestinal commensals present in poultry at different ages (Amit-Romach et al., 2004; Skansenga et al., 2006).
In positive control sample, an unambiguous band corresponding to a bacterial chromosome on agarose gels was established. Subsequent PCRs performed with negative samples did not show any amplification. However, 722bp fragments were amplified when DNA from a Clostridium sp.-positive control was added to the negative samples (Fig 1). The robustness of the method was based on positive and negative control samples processed throughout the entire procedure.
DISCUSSION
Conventional methods for Clostridium sp. detection by the microbiological, serological, biochemical and other approaches present serious difficulties for standard selection. There is no general agreement concerning the determination of the gold standard for the detection of this foodborne pathogen. To date, culture techniques are universally recognized as the standard method for the detection of bacterial groups such as Clostridium sp. in broilers gastrointestinal. In theory, these methods are capable of detecting as few as one viable cell in a sample following pre- and selective enrichment stages. However, increased sensitivity of PCR methods, compared to culture techniques, has been reported for the detection of Clostridium sp. (Engstrom et al., 2003; Gholamiandekhordi et al. 2006) and has been attributed to the fact that PCR could detect the target sequences, irrespective of the growth potential, of target cells.
The method presented in this work was developed for routine detection of a large number of samples on a daily basis. Meanwhile, the PCR mixture was prepared in a large volume, aliquoted into PCR 0.2ml micro-tubes. Furthermore, results showed the rapid detection of Clostridium sp. by simplifying the entire detection process, since the entire detection process took less than half day, showing that the time needed to diagnose was greatly reduced.
In the future, a multiplex PCR test could be developed, which would allow the analysis of a complex microflora in a single or a limited number of reactions, taking advantage of the rapidly growing number of 16S RNA sequences available.
The analysis of the microbial contents of the different small intestine segments examined indicated that Clostridium sp. was not consistently detected as same in all the intestinal segments. In fact, posterior segments exhibited higher levels of Clostridium sp at 4 and 30 d. compared with the anterior segments especially cecum.
The PCR protocol used in this work detected Clostridium sp. in naturally contaminated intestinal samples efficiently. The incidence of Clostridium sp. in poultry gut samples was high, but similar to that reported by authors from other countries such as Johansson et al. (2004), Olkowski et al. (2006) etc. These high incidences were not surprising if the spread of microorganisms in environment, diet, water, litter, slaughtering, etc were considered. The detection of Clostridium sp. in live birds could be a result of contaminated feed. Harris et al (1997) reported that animal and poultry feeds were sometimes contaminated with pathogenic bacteria. Contaminated water could also be another medium of transmission.
The present results improved the efficacy of detection of Clostridium sp in intestinal samples at different ages. In the present study, duodenum, jejunum, ileum and cecum samples were collected from 24 chickens. Since the samples were from the similar chickens, the bacterial populations represented the gut microflora in the duodenum, jejunum, ileum and cecum of these birds as a whole, regardless of differences in microflora of individual chickens. It is known that the gut microflora cans significantly be influenced by the diets and other factors, such as the hosts and environment. Therefore, the data presented in this study should be considered to be case-specific (Gong et al., 2002).
The present study has also revealed the heterogeneity of Clostridium sp. present in the duodenum, jejunum, ileum and cecum. The differences of Clostridium sp. existence in these four segments likely resulted from the interactions of different animal host tissues/cells and gut microflora. The function of the ileum (the lower end of the small intestine) is mainly nutrient absorption, while the cecum is the site where extensive bacterial fermentation occurs, resulting in further nutrient absorption and detoxification of substances that are harmful to the host (Csordas, 1995). Since these segments function differently and provide different environments, it is expected that different types of bacteria would colonize them and distinct microflora would develop (Gong et al., 2002). In this study, some differences of the Clostridium sp. was demonstrated in the four intestinal segments.
ACKNOWLEDGEMENTS
The excellent technical assistance of Dr Ali Qotbi and Payam Potki is greatly appreciated. We thank Alireza Bizhannia for helping in collecting broiler samples. We also thank the Agriculture Biotechnology Research Institute of Iran for financial support.
Received: September 05, 2007; Revised: December 18, 2008; Accepted: June 09, 2009.
- Amit-Romach, E.; Sklan, D. and Uni, Z. (2004), Microflora Ecology of the Chicken Intestine Using 16S Ribosomal DNA Primers. Poult. Sci 83, 1093-1098.
- Apajalahti, J.H.; Sarkilahti, L.K.; Maki, B.R.; Heikkinen, J.P.; Nurminen, P.H. and Holben, W.E. (1998), Effective recovery of bacterial DNA and percent guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl Environ Microbiol 64, 4084-4088.
- Barbara, A.J.; Trinh, H.T.; Glock, R.D. and Songer, J.G. (2007), Necrotic enteritis-producing strains of Clostridium perfringens displace nonnecrotic enteritis strains from the gut of chicks. Veterinary Microbiology (In Press). doi:10.1016/j. vetmic. 2007.07.019.
- Carlin, F.; Broussolle, V.; Perelle, S.; Litman, S. and Fach, P. (2004),. Prevalence of Clostridium botulinum in food raw materials used in REPFEDs manufactured in France. Int. J. Food Microbiol 91, 141-145.
- Csordas, A. (1995), Toxicology of butyrate and short-chain fatty acids. In: Role of Gut Bacteria in Human Toxicology and Pharmacology (Hill, M.J., Ed.), pp. 105-125. Taylor and Francis, London.
- Engström, B.E.; Fermér, C.; Lindberg, A.; Saarinen, E.; Båverud, V. and Gunnarsson, A. (2003), Molecular typing of isolates of Clostridium perfringens from healthy and diseased poultry. Vet. Microbiol 94, 225-235.
- Ficken, M.D. and Wages, D.P. (1997), Necrotic enteritis. In: Barnes, H.J.; Beard, C.W.; McDougald, L.; Saif, Y.M. and Calnek, B.W. (Eds.), Diseases of Poultry. Iowa State University press, Ames, IA, pp. 261-264.
- Gholamiandekhordi, A.R.; Ducatelle, R.; Heyndrickx, M.; Haesebrouck, F. and Van-Immerseel, P. (2006), Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet. Microbiol 113, 143-152.
- Gong, J.; Forster, R.J.; Yu, H.; Chambers, J.R.; Wheatcroft, R.; Sabour, P.M. and Chen, S. (2002), Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol 41, 171-179.
- Grave, K.; Kaldhusdal, M.; Kruse, H.; FevangHarr, L.M. and Flatlandsmo, K. (2004), What has happened in Norway after the ban of Avoparcin? Consumption of antimicrobials by poultry. Preventive Vet. Med 62, 59-72.
- Harris, I.T.; Fedorka-Caray, P.J.; Gray, J.T.; Thomas, L.A. and Ferris, K. (1997), Prevalence of Salmonella organisms in swine feed. J. American Vet. Med. Assoc 1210(3), 382-385.
- Hill, J.E.; Seipp, R.P.; Betts, M.; Hawkins, L.; Van Kessel, A.G. and Crosby, W.L. (2002), Extensive profiling of a complex microbial community by highthroughput sequencing. Appl. Environ. Microbiol 68, 3055-3066.
- Jaimes, C.P.; Aristizábal, F.A.; Bernal, M.; Suárez, Z.R. and Montoya, D. (2006), AFLP fingerprinting of Colombian Clostridium spp. strains, multivariate data analysis and its taxonomical implications. J. Microbiol. Methods. 67, 64-69.
- Johansson, A.; Greko, C.; Engström, B.E. and Karlsson, M. (2004), Antimicrobial susceptibility of Swedish, Norwegian and Danish isolates of Clostridium perfringens from poultry, and distribution of tetracycline resistance genes. Vet. Microbiol 99, 251-257.
- Kaldhusdal, M.; Schneitz, C.; Hofshagen, M. and Skjerve, E. (2001), Reduced incidence of Clostridium perfringens-associated lesions and improved performance in broiler chickens treated with normal intestinal bacteria from adult fowl. Avian Dis 45, 149-156.
- La-Ragione, R.M. and Woodward, M.J. (2003), Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet. Microbiol 94, 245-256.
- Lu, J.; Domingo, J.S. and Shanks, O.C. (2007), Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res 41, 3561-3574.
- Lu, J.; Idris; J.; Harmon, B.G.; Hofacre, C.; Maurer, J.J. and Lee, M.D. (2003), Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol 69, 6816-6824.
- McCourt, M.T.; Finlay, D.A.; Laird, C.; Smyth, J.A.; Bell, C. and Ball, H.J. (2005); Sandwich ELISA detection of Clostridium perfringens cells and α-toxin from field cases of necrotic enteritis of poultry. Vet. Microbiol 106, 259-264.
- National Research Council (NRC). (1994), Nutrient Requirement of Poultry. 9th ed. National Academy. Washington DC.
- Nauerby, B.; Pedersen, K. and Madsen, M. (2003), Analysis by pulsed-field gel electrophoresis of the genetic diversity among Clostridium perfringens isolates from chickens. Vet. Microbiol. 94, 257-266.
- Olkowski, A.A.; Wojnarowicz, C.; Chirino-Trejo, M. and Drew, M.D. (2006), Responses of broiler chickens orally challenged with Clostridium perfringens isolated from field cases of necrotic enteritis. Res in Vet Sci 81, 99-108.
- Petit, L.; Gibert, M. and Popoff, M.R. (1999), Clostridium perfringens: toxinotype and genotype. Trends Microbiol 7, 104-110.
- Prescott, J.F.; Sivendra, R. and Barnum, D.A. (1978), The use of bacitracin in the prevention and treatment of experimentallyinduced ecrotic enteritis in the chicken. Can. Vet. J 19, 181-183.
- Seidavi, A.R.; Mirhosseini, S.Z.; Shivazad, M.; Chamani, M.; Sadeghi, A.A. and Pourseify, R. (2007), Effect of five different components on optimization of PCR for detection of Clostridium spp. bacteria in broiler gastrointestinal tract. Vet. J. Islamic Azad University, Garmsar Branch 3, 153-162.
- Shanmugavelu, S.; Ruzickova, G.; Zrustova, J. and Brooker, J.D. (2006), A fermentation assay to evaluate the effectiveness of antimicrobial agents on gut microflora. J. Microbiol Methods 67, 93-101.
- Skansenga, B.; Kaldhusdalc, M. and Rudi, K. (2006), Comparison of chicken gut colonisation by the pathogens Campylobacter jejuni and Clostridium perfringens by real-time quantitative PCR. Mol and Cell Probes 20, 269-279.
- Tech, R. (1999), Necrotic enteritis and associated conditions in broiler chickens. World Poultry. 15, 44-47.
- Titball, R.W. (1993), Bacterial phospholipases C. Microbiol. Rev 57, 347-366.
- Zhu, X.Y.; Zhong, T.; Panya, Y. and Joerger, R.D. (2002), 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol 68, 124-137.
- Zoetendal, E.G.; Collier, C.T.; Koike, S.; Mackie, R.I. and Gaskins, H.P. (2004), Molecular Ecological Analysis of the Gastrointestinal Microbiota: A Review. The J. of Nutrition 134, 465-472.
Publication Dates
-
Publication in this collection
25 Feb 2010 -
Date of issue
Feb 2010
History
-
Accepted
06 Sept 2009 -
Reviewed
18 Dec 2008 -
Received
05 Sept 2007