Abstracts
Many viral diseases, including leafroll, which is of great economic importance, affect grapevines (Vitis spp.). A complex of eight viruses [Grapevine leafroll-associated virus (GLRaV) -1 to 8] is associated with this disease. The objective of this study was to compare the variability of the 3' terminal region of the polymerase gene of three isolates of GLRaV-3 (Grapevine leafroll-associated virus-3), from Submédio do Vale do Rio São Francisco (Petrolina-PE) with that of other isolates available at the GenBank, including an isolate from North America and another from Southern Brazil. The viral RNA was extracted from three infected ELISA reactive plants and a fragment of 340 bp was amplified, by RT-PCR, using primers that recognize that portion of the polymerase gene found between nucleotides 8267 and 8606. The three isolates from Vale do Rio São Francisco named Pet-1, Pet-2 and Pet-3, showed similarities ranging from 98% and 94%, respectively to the isolates from North America (AF037268) and Southern Brazilian (AF438411). Considering the whole genome, the main variation found was one amino acid change at position 2766 (F2766Y). These preliminary data indicate the existence of a natural variation among GLRaV-3 isolates from grapevines. This could be due to the vegetative propagation and long cycle of the plant, associated with the error-prone nature of RNA-dependent RNA polymerase.
GLRaV-3; Vitis spp.; Ampelovirus; RNA dependent RNA polymerase
A videira (Vitis spp.) é afetada por diversas viroses, dentre essas, o enrolamento da folha se destaca pela importância econômica. A doença é causada por um complexo formado por até oito vírus [Grapevine leafroll-associated virus (GLRaV) -1 ao -8]. O objetivo desse trabalho foi comparar a variabilidade da extremidade 3' do gene da polimerase de isolados de GLRaV-3, provenientes do Submédio do Vale do Rio São Francisco (Petrolina, PE) com a de outros isolados disponíveis no GenBank, incluindo um isolado da América do Norte e um da região sul do Brasil. O RNA viral foi extraído de três amostras infetadas, reagentes em teste de ELISA, e um fragmento de 340 pb foi amplificado por RT-PCR, utilizando-se oligonucleotídeos para a região do gene da polimerase viral compreendida entre os nucleotídeos 8267 a 8606. Observou-se que os três isolados da região do São Francisco, denominados Pet-1, Pet-2 e Pet-3, apresentaram 98% e 94% de similaridade com o isolado norte-americano (AF037268) com aquele do sul do Brasil (AF438411), respectivamente. A principal variação observada foi uma troca de um aminoácido da posição 2766 (F2766Y), considerando-se o genoma completo. Esses dados preliminares indicam a existência de uma variabilidade natural entre isolados de GLRaV-3 em videiras. Isso pode ser explicado pela propagação vegetativa e pelo longo ciclo da planta associados à propensão ao erro da RNA polimerase dependente de RNA.
GLRaV-3; Vitis spp.; Ampelovirus; RNA dependent RNA polymerase
COMMUNICATIONS COMUNICAÇÕES
Variability of the 3' terminal of the polymerase gene of Grapevine leafroll-associated virus 3 isolates from Vale do São Francisco, Brazil* * Part of the Master Dissertation of the first author. Universidade de Brasília (2004). ** CAPES fellowship
Variabilidade da extremidade 3' do gene da polimerase de isolados de Grapevine leafroll-associated virus-3 do Submédio do Vale do São Francisco
Érico de C. DianeseI, ** * Part of the Master Dissertation of the first author. Universidade de Brasília (2004). ** CAPES fellowship ; Eduardo D. RamalhoI; Daniela M. CerqueiraI; Daniela B. LopesIII; Thor V.M. FajardoII; Marisa A.S.V. FerreiraI; Cláudia R.F. MartinsI
IDepartamento de Fitopatologia, Instituto de Biologia, Universidade de Brasília, Campus UnB, CEP 70910-900, Brasília, DF, fax: (061)272-1793
IIEmbrapa Uva e Vinho, Cx. Postal 130, CEP 95700-000, Bento Gonçalves, RS
IIIEmbrapa Semi-Árido, Cx. Postal 23, CEP 56300-000, Petrolina, PE, e-mail: cmartins@unb.br
ABSTRACT
Many viral diseases, including leafroll, which is of great economic importance, affect grapevines (Vitis spp.). A complex of eight viruses [Grapevine leafroll-associated virus (GLRaV) -1 to 8] is associated with this disease. The objective of this study was to compare the variability of the 3' terminal region of the polymerase gene of three isolates of GLRaV-3 (Grapevine leafroll-associated virus-3), from Submédio do Vale do Rio São Francisco (Petrolina-PE) with that of other isolates available at the GenBank, including an isolate from North America and another from Southern Brazil. The viral RNA was extracted from three infected ELISA reactive plants and a fragment of 340 bp was amplified, by RT-PCR, using primers that recognize that portion of the polymerase gene found between nucleotides 8267 and 8606. The three isolates from Vale do Rio São Francisco named Pet-1, Pet-2 and Pet-3, showed similarities ranging from 98% and 94%, respectively to the isolates from North America (AF037268) and Southern Brazilian (AF438411). Considering the whole genome, the main variation found was one amino acid change at position 2766 (F2766Y). These preliminary data indicate the existence of a natural variation among GLRaV-3 isolates from grapevines. This could be due to the vegetative propagation and long cycle of the plant, associated with the error-prone nature of RNA-dependent RNA polymerase.
Additional keywords: GLRaV-3, Vitis spp., Ampelovirus, RNA dependent RNA polymerase.
RESUMO
A videira (Vitis spp.) é afetada por diversas viroses, dentre essas, o enrolamento da folha se destaca pela importância econômica. A doença é causada por um complexo formado por até oito vírus [Grapevine leafroll-associated virus (GLRaV) -1 ao -8]. O objetivo desse trabalho foi comparar a variabilidade da extremidade 3' do gene da polimerase de isolados de GLRaV-3, provenientes do Submédio do Vale do Rio São Francisco (Petrolina, PE) com a de outros isolados disponíveis no GenBank, incluindo um isolado da América do Norte e um da região sul do Brasil. O RNA viral foi extraído de três amostras infetadas, reagentes em teste de ELISA, e um fragmento de 340 pb foi amplificado por RT-PCR, utilizando-se oligonucleotídeos para a região do gene da polimerase viral compreendida entre os nucleotídeos 8267 a 8606. Observou-se que os três isolados da região do São Francisco, denominados Pet-1, Pet-2 e Pet-3, apresentaram 98% e 94% de similaridade com o isolado norte-americano (AF037268) com aquele do sul do Brasil (AF438411), respectivamente. A principal variação observada foi uma troca de um aminoácido da posição 2766 (F2766Y), considerando-se o genoma completo. Esses dados preliminares indicam a existência de uma variabilidade natural entre isolados de GLRaV-3 em videiras. Isso pode ser explicado pela propagação vegetativa e pelo longo ciclo da planta associados à propensão ao erro da RNA polimerase dependente de RNA.
Palavras-chave adicionais: GLRaV-3, Vitis spp., Ampelovirus, RNA dependent RNA polymerase.
Grapevines (Vitis spp.) are affected by a great number of viruses that can diminish productivity, compromise the quality of the grapes and, in some cultivars, cause plant decay. In Brazil, grapevine leafroll stands out as an economically important disease caused by viruses. An incidence of 78% has been reported in São Paulo, and one of 15.6 - 98% in Rio Grande do Sul. It has also been found and described in the States of Goiás, Minas Gerais, Paraná, Santa Catarina and Bahia/Pernambuco (Vale do São Francisco) (Tavares et al., 2000). Severely affected plants have been observed to suffer a reduction of 42.4% in number of clusters, 62.8% in production and 65.2% in vigor (Kuhn & Nickel, 1998; Fajardo et al., 2002).
The complex responsible for this disease is composed of eight virus species named Grapevine leafroll-associated virus-1 to 8 (GLRaV-1 to 8) (Martelli et al., 2002). The viruses are transmitted in the field by grafting and through pseudococcid (Pseudococcus sp. and Planococcus sp.) and coccid from the genera Pulvinaria, Neopulvinaria and Parthenolecanium, mealybugs. Vector transmission has been confirmed for two viral species of the complex (GLRaV-1 and -3); field transmission has also been confirmed in each if these (Fortusini et al., 1996; Habili et al., 1995).
With the exception of GLRaV-2 and GLRaV-7, all other GLRaV species belong to the family Closteroviridae, genus Ampelovirus, and the type specie of the genus is (Grapevine leafroll associated virus -3) (Martelli et al, 2002). The viral particles are filamentous with 1,500 to 2,200 nm in length, presenting a 15 to 20 kb single stranded RNA genome and a capsid protein molecular weight ranging from 35 to 43 kDa (the GLRaV-2 capsid protein has 26 kDa) (Zimmermann et al., 1990; Martelli et al., 2002).
In the field, visual confirmation of the Vitis vinifera L. infection is relatively easy. During the spring, the leaves of infected and healthy plants are quite similar. However, throughout the other seasons, the infected leaves become yellow or red, depending on the cultivar, and the leaves roll down (Goheen, 1988). The most common symptom on grapevine clusters, seen mainly in red vineyards, is the irregular or retarded maturation of the grapes. In some cases, when the plant is severely affected, the maturation process is completely compromised, coming to a halt. American cultivars of Vitis labrusca L., hybrids and rootstocks exhibit very mild or no leafroll symptom (Kuhn & Nickel, 1998).
The study of the viral complex is very difficult, due to particular characteristics shared by all members of the Closteroviridae family. One of these special characteristics is the restriction of the virus particles to phloem tissues. This makes the purification processes very difficult, resulting in insufficient yields for in-depth analysis. Two other factors that also make it more difficult to study GLRaV are: 1) the viruses of the genus Ampelovirus have a long genomic RNA molecule; 2) they are not mechanically transmitted (Karasev, 2000).
The objective of this work was to compare the variability of a fragment of the viral polymerase gene of GLRaV-3 isolated from grapevines grown in the Vale do São Francisco, Pernambuco, Brazil, to two other sequences available at the GenBank, one from a North American isolate (AF037268) and the other an isolate from Southern Brazilian (AF438411). This segment was chosen because it is the only genomic region of GLRaV-3 with more than one sequence available at the GenBank.
Initially, the enzyme linked immunosorbent assay (ELISA) (Sanofi Diagnostic Pasteur) was performed to detect GLRaV-3 infected plants in samples from the Alicante Bouchet grapevine cultivar in the Vale do São Francisco. A total of 16 samples from different plants collected within a period of six months were tested. None of them showed any biological difference in symptoms. Three plants presented reactive results in ELISA and were submitted to RNA extraction and PCR. Characteristic leafroll symptom and dark colored internerval regions were observed on the tested plants. These symptomatic plants were maintained in a greenhouse at the Estação Experimental da Biologia, Instituto de Ciências Biológicas, Universidade de Brasília.
The RNA extraction was performed using the method described by Mackenzie (1997), with modifications. A total of 0.1 g of leaf tissue (including petioles) was ground in liquid nitrogen. Lysis buffer was added (0.2 M sodium acetate, pH 5.0, 25 mM EDTA, 2.5% (wt/vol) PVP-40, and 1% (vol/vol) 2-mercaptoethanol) and the tissue was ground again. A centrifugation cycle was performed at 3,000 g for 5 min. The supernatant was transferred to another tube and the QIAamp RNA extraction kit protocol (Qiagen) was used.
The reverse transcription reaction was made in a final volume of 20 µl (200 U of reverse transcriptase, 150 mg of random primers and 7 µl of extraction reaction). The amplification of the 340 bp fragment of the viral polymerase was made possible by the use of the primers C547 and H229 (Minafra & Hadidi, 1994) and the program proposed by Mackenzie et al. (1997) with modifications: 95 ºC for 2 min following 35 cycles of denaturation at 95 ºC for 1 min, annealing at 53 ºC for 1 min, elongation at 72 ºC for 1 min and final elongation at 72 ºC for 7 min. The amplified fragments were purified, using a commercial kit (Amersham-Pharmacia), and cloned into pGEM- T Easy vector (Promega).
Two different PCR products of each of the three cv. Alicante Bouchet infected plants were cloned and sequenced in both directions (forward and reverse), in a Megabace System sequencer (Amersham-Pharmacia). The generated sequences contained 340 bp each. The three sequences derived from different plants were compared to two other sequences available at the GenBank (AF037268 and AF438411). This comparison was done using the programs BLAST and Bioedit (Altschul et al., 1990; Hall, 1999).
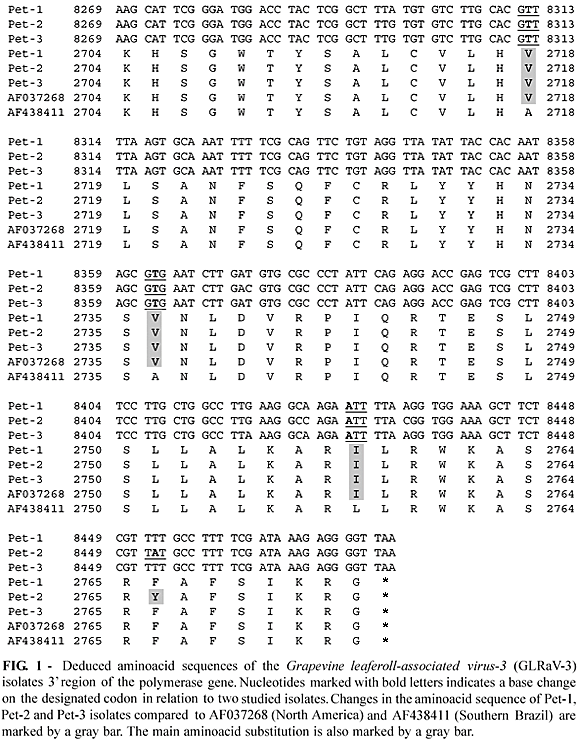
The three samples showed nucleotide substitutions when compared to each other. (Figure 1). The nucleotide sequence of the first and second isolates (Pet-1 and Pet-2) demonstrated an identity of 98% and 94%, respectively to the North American (AF037268) and Southern Brazilian isolates (AF438411). The third isolate presented a polymorphism at position 8418. Two clones had an adenine and two a guanine in that position. When adenine was present, the third isolate (Pet-3) showed an identity of 98% and 96%, respectively to isolates NY1 and AF438411. When guanine was present, it showed an identity of 99% and 96%. These substitutions were observed as two possible viral populations infecting the same plant, resulting in slight changes in the sequence. The nucleotide sequences of the three isolates were translated and the deduced aminoacid sequences were compared. Pet-1 and Pet-3 showed an identity of 100% with AF037268 and 95% while AF438411. Pet-2 showed an identity of 98% and 94% respectively (Figure 1).
The results obtained through the comparison of the three generated sequences (Pet-1, Pet-2 and Pet-3) with the Brazilian isolate AF438411 showed amino acid differences of 4 to 6%. This indicates a variability that could be explained as a natural variation of GLRaV-3 isolates within grapevines, it could also be due to the vegetative propagation and long cycle of the plant, associated with error prone RNA-dependent RNA polymerase. The variation may also be influenced by the plant-pathogen interaction responsible for originating viral variants (Little et al., 2001).
An observation of the deduced aminoacid sequence showed that the differences in nucleotide sequences were of little influence on the protein product. The aminoacid substitutions were mainly conservative. The main variation found was the aminoacid substitution at position 2766 (F2766Y) of the whole genome when comparing Pet-2 and NY1 (Figure 1). This work represents a joint effort of three Brazilian institutions working towards a deeper understanding of the genetic variability of viruses affecting grapevines. Very little is known about the natural variability of GLRaV-3, since only one isolate has been totally sequenced at this time.
ACKNOWLEDGEMENTS
This project was supported by Banco do Nordeste do Brasil, project nº 1616/2000, FINATEC and Conselho Nacional de Desenvolvimento Científico (CNPq). We thank John Penney, for reviewing this manuscript.
LITERATURE CITED
Accepted for publication on 20/12/2005
Corresponding author: Cláudia R. F. Martins
- ALTSCHUL, S.F., GISH, W., MILLER, W., MYERS, E.W. & LIPMAN, D.J. Basic local alignment search tool. Journal of Molecular Biology 215:403-410. 1990.
- FAJARDO, T.V.M., KUHN, G.B., EIRAS, M. & NICKEL, O. Detecção de Closterovirus em videira e caracterização parcial de um isolado do Grapevine leafroll-associated virus 3. Fitopatologia Brasileira 27:58-64. 2002.
- FORTUSINI, A., SCATTINI, G., CINQUANTA, S. & PRATI, S. Diffusione naturale del virus 1 (GLRaV-1) del virus 3 (GLRaV-3) dell' accartociamento fogliare e del virus della maculatura infettiva o "fleck" (GFkV) della vite. Informatore Fitopatologico 12:39-43. 1996.
- GOHEEN, A.C. Leafroll. In: Pearson, R.C. & Goheen, A.C. Compendium of grape diseases. APS Press, St. Paul. 1988. pP.52.
- HABILI, N., FAZELI, C.F., EWART, A., HAMILTON, R., CIRAMI, R., SALDARELLI, P., MINAFRA, A. & REZAIAN, M.A. Natural spread and molecular analysis of grapevine leafroll-associated virus 3 in Australia. Phytopathology 85:1418-1422. 1995.
- HALL, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95-98. 1999.
- KARASEV, A.V. Genetic diversity and evolution of closteroviruses. Annual Review of Phytopathology 38:293-324. 2000.
- KUHN, G.B. & NICKEL, O. Viroses e sua importância na viticultura brasileira. Informe Agropecuário 19:85-91.1998.
- LING, K.S., ZHU, H.Y., DRONG, R.F., SLIGHTOM, J.L., MCFERSON, J.R. & GONSALVES, D. Nucleotide sequence of the 3'-terminal two-thirds of the grapevine leafroll-associated virus-3 genome reveals a typical monopartite closterovirus. Journal of General Virology 79:1299-1307. 1998.
- LITTLE, A., FAZELI, C.F., REZAIAN, M.A. Hypervariable genes in grapevine leafroll associated virus 1. Virus Research 80:109-116. 2001.
- MACKENZIE, D.J., MCLEAN, M.A., MUKERJI, S. & GREEN, M. Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Disease 81:222-226. 1997.
- MARTELLI, G.P., AGRANOVSKY, A.A., BAR-JOSEPH, M., BOSCIA, D., CANDRESSE, T., COUTTS, R.H.A., DOLJA, V.V., FALK, B.W., GONSALVES, D., JELKMANN, W., KARASEV, A.V., MINAFRA, A., NAMBA, S., VETTEN, H.J., WISLER, G.C., YOSHIKAWA, N. The family Closteroviridae revised. Archives of Virology. 147:2039-2044. 2002.
- MINAFRA, A. & HADIDI, A. Sensitive detection of grapevine virus A, B, or leafroll-associated III from viruliferous mealybugs and infected tissue by cDNA amplification. Journal of Virological Methods 47:175-188. 1994.
- TAVARES, S.C.C.H., LIMA, M.F. & MELE, N.F. Principais doenças da videira e alternativas de controle. In: A Viticultura no Semi-árido Brasileiro. Embrapa Semi-árido. 2000. pp. 318-339.
- ZIMMERMANN, D., BASS, P., LEGIN, R. & WALTER, B. Characterization and serological detection of four closterovirus like particles associated with leafroll disease on grapevine. Journal of Phytopathology 130:205-218. 1990.
Publication Dates
-
Publication in this collection
17 May 2005 -
Date of issue
Apr 2005
History
-
Accepted
20 Dec 2005