Abstracts
Fungal diseases in cotton (Gossypium hirsutum), such as anthracnose caused by Colletotrichum gossypii and ramulose caused by C. gossypii var. cephalosporioides, are responsible for large yield losses. These pathogens are seed borne and morphologically similar although they induce different symptoms, which can lead to misdiagnosis using the blotter testing method. The present study was carried out to assess the viability of using Amplified Fragment Length Polymorphism (AFLP) markers to differentiate these pathogens. Five isolates, for each pathogen, were classified according to pathogenicity on cotton plants, and mycelial growth morphology. Conidial suspensions were sprayed on 30-day-old cotton plants and the symptoms assessed ten and 40 days after inoculation. For growth morphology 200 cottonseeds were inoculated with seven-day-old pure cultures, and the mycelial traits observed under a stereoscopic microscope seven days after inoculation. The DNA for AFLP analysis was obtained from seven-day-old fungal mycelia grown in liquid medium, using the Dneasy Qiagen protocol. Using the AFLP technique 318 polymorphic bands were selected to estimate similarities using Dice's Coefficient. The results clearly distinguished between ramulose and anthracnose isolates, which agreed with morphological and pathogenicity testing.
molecular markers; anthracnose; ramulose; cotton diseases; seed borne pathogens
Doenças fúngicas em algodoeiro (Gossypium hirsutum), tais como antracnose causada por Colletotrichum gossypii e ramulose causada por C. gossypii var. cephalosporioides, levam a grandes perdas em produtividade. Estes microrganismos são patógenos veiculados por sementes e morfologicamente similares apesar deles induzirem sintomas diferentes, que podem levar a um erro no diagnóstico usando o teste de sanidade com sementes em rolo de papel. O presente estudo foi realizado para acessar a viabilidade do uso de marcadores Amplified Fragment Length Polymorphism (AFLP) para diferenciar estes patógenos. Cinco isolados, para cada patógeno, foram classificados de acordo com a patogenicidade e morfologia de crescimento micelial. As suspensões de conídios foram pulverizadas em plantas de algodoeiro com 30 dias e os sintomas avaliados aos dez e 40 dias após a inoculação. Para a morfologia de crescimento micelial, 200 sementes foram inoculadas com culturas puras com sete dias, e as características miceliais observadas sob microscópio esterioscópico sete dias após a inoculação. O DNA para a análise de AFLP foi obtido de micélio fúngico com sete dias, crescido em meio líquido, usando o protocolo Dneasy Qiagen. A técnica AFLP produziu 318 bandas polimórficas que foram selecionadas para calcular as similaridades usando o Coeficiente de Dice. Os resultados claramente distinguiram os isolados causadores de ramulose dos de antracnose, os quais concordam, com dados morfológicos e de patogenicidade.
marcadores moleculares; antracnose; ramulose; doenças do algodoeiro; patógenos de sementes
COMMUNICATIONS COMUNICAÇÕES
AFLP markers differentiate isolates of Colletotrichum gossypii from C. gossypii var. cephalosporioides* * Part of the Doctor Thesis of the first author. Universidade Federal de Lavras. (2002). ** CAPES fellowship
Marcadores AFLP diferenciam Colletotrichum gossypii de C. gossypii var. cephalosporioides
Renata Silva-MannI, ** * Part of the Doctor Thesis of the first author. Universidade Federal de Lavras. (2002). ** CAPES fellowship ; Maria G. G. Carvalho VieiraII; José C. MachadoIII; José Roberto Bernardino FilhoII; Kalinka C. C. SalgadoII; Mikel R. StevensIV
IDepartamento de Eng. Agronômica, Universidade Federal de Sergipe, Cidade Universitária Prof. José Aloísio de Campos, Av. Marechal Rondon s/n, Jardim Rosa Elze, CEP 49100-000, São Cristóvão, SE, e-mail: renatamann@hotmail.com
IILaboratório de Análise de Sementes-DAG/UFLA, e-mail: semente@ufla.br
IIILaboratório de Patologia de Sementes-DFS/UFLA, e-mail: machado@ufla.br, Cx. Postal 37, CEP 37200-000, Lavras, MG
IVDepartment of Plant and Animal Sciences, Brigham Young University, Provo, UT, USA e-mail: mikel_stevens@byu.edu
ABSTRACT
Fungal diseases in cotton (Gossypium hirsutum), such as anthracnose caused by Colletotrichum gossypii and ramulose caused by C. gossypii var. cephalosporioides, are responsible for large yield losses. These pathogens are seed borne and morphologically similar although they induce different symptoms, which can lead to misdiagnosis using the blotter testing method. The present study was carried out to assess the viability of using Amplified Fragment Length Polymorphism (AFLP) markers to differentiate these pathogens. Five isolates, for each pathogen, were classified according to pathogenicity on cotton plants, and mycelial growth morphology. Conidial suspensions were sprayed on 30-day-old cotton plants and the symptoms assessed ten and 40 days after inoculation. For growth morphology 200 cottonseeds were inoculated with seven-day-old pure cultures, and the mycelial traits observed under a stereoscopic microscope seven days after inoculation. The DNA for AFLP analysis was obtained from seven-day-old fungal mycelia grown in liquid medium, using the Dneasy Qiagen protocol. Using the AFLP technique 318 polymorphic bands were selected to estimate similarities using Dice's Coefficient. The results clearly distinguished between ramulose and anthracnose isolates, which agreed with morphological and pathogenicity testing.
Additional keywords: molecular markers, anthracnose, ramulose, cotton diseases, seed borne pathogens.
RESUMO
Doenças fúngicas em algodoeiro (Gossypium hirsutum), tais como antracnose causada por Colletotrichum gossypii e ramulose causada por C. gossypii var. cephalosporioides, levam a grandes perdas em produtividade. Estes microrganismos são patógenos veiculados por sementes e morfologicamente similares apesar deles induzirem sintomas diferentes, que podem levar a um erro no diagnóstico usando o teste de sanidade com sementes em rolo de papel. O presente estudo foi realizado para acessar a viabilidade do uso de marcadores Amplified Fragment Length Polymorphism (AFLP) para diferenciar estes patógenos. Cinco isolados, para cada patógeno, foram classificados de acordo com a patogenicidade e morfologia de crescimento micelial. As suspensões de conídios foram pulverizadas em plantas de algodoeiro com 30 dias e os sintomas avaliados aos dez e 40 dias após a inoculação. Para a morfologia de crescimento micelial, 200 sementes foram inoculadas com culturas puras com sete dias, e as características miceliais observadas sob microscópio esterioscópico sete dias após a inoculação. O DNA para a análise de AFLP foi obtido de micélio fúngico com sete dias, crescido em meio líquido, usando o protocolo Dneasy Qiagen. A técnica AFLP produziu 318 bandas polimórficas que foram selecionadas para calcular as similaridades usando o Coeficiente de Dice. Os resultados claramente distinguiram os isolados causadores de ramulose dos de antracnose, os quais concordam, com dados morfológicos e de patogenicidade.
Additional keywords: marcadores moleculares, antracnose, ramulose, doenças do algodoeiro, patógenos de sementes.
Fungal infections account for large crop losses, especially in the Brazilian rainy season. Studies have identified a number of fungal species associated with cotton (Gossypium hirsutum L.) seeds in Brazil, along with the description of the effects of pathogenic fungi on seed germination and disease transmission to virgin cotton lands (Teixeira, 2001).
Anthracnose caused by Colletotrichum gossypii South, causes lesions on all parts of the cotton plant, while ramulose is caused by C. gossypii (South) var. cephalosporioides A. S. Costa is the most severe cotton disease in Brazil (Davis et al., 1981). Anthracnose is mainly a seedling disease that causes "damping off". The two pathogens cannot be distinguished using the paper-blotting test. Therefore, this research was conducted to assess the viability of utilizing the Amplified Fragment Length Polymorphism (AFLP) marker technique (Zabeau, 1993) to distinguish C. gossypii from C. gossypii var. cephalosporioides.
Inoculum was prepared by growing Colletotrichum spp. isolates (Table 1) on potato-dextrose-agar (PDA) medium for seven days. Inoculations were carried out by spraying 30-day-old plants of IAC-22, ITA-90 (CNPA Itamarati 90), ITA-96 (CNPA Itamarati 96), Redenção (Epamig-4) and Precoce (Epamig-5) cultivars with a 106 conidia/ml suspension. Sterilized de-ionized water was utilized as a control. Inoculated plants were kept in a moist chamber for 72 h. Disease assessment was carried out ten and 40 days after inoculation, following a modified version of the disease severity scale of Cia (1977) for anthracnose and ramulose, as follows: (1) Plant without symptoms (no lesions); (2) Plant with star spots on the top leaves; (3) Plant with shortened internodes (to as low as 60% of the control) and with circle and star leaf spots on the top leaves; (4) Plant with over sprouting and reduced internodes of 40 to 60% of the controls; (5) Plant with over sprouting and reduced growth, reduced internodes less than 60% of the control.
Cottonseeds (cultivars IAC-22 e ITA-90) were inoculated on seven-day-old pure cultures of C. gossypii and C. gossypii var. cephalosporioides. Seven days after inoculation the blotter test (Brasil, 1992) was performed using a random experimental design with four replications of 50 seeds each, and the mycelial traits observed under a stereoscopic microscopy (Tanaka et al., 1996).
For AFLP analysis, mycelia from monosporic cultures were obtained by cultivating isolates in liquid medium (10 g glucose; 1.0 g de NH4H2PO4; 0.2 g KCl; 0.2 g MgSO4. 7 H2O; 5 g yeast extract; 1 ml CuSO4. 5 H2O; 5% polyvinyl-pyrrolidone; 1 ml ZnSO4: 7 H2O. 1% p/v for 1000 ml sterile distilled water) (Mills et al., 1994) for seven days at 22ºC in orbital shaker (Model MA 140) at 170 rpm. The mycelia were filtered and lyophilized for 48 h at -40 ºC and 10-1 mbar pressure. Total DNA extraction was performed using the DNeasy Qiagen Kit.
Digestion of genomic DNA was performed in 1.5 ml tube with 2.5 ìl of DNA (10ng/ìl), 5.0 ìl of 5X buffer reaction (Life Technologies Gibco BRL), 4 U/µl of MseI, 10 U/µl EcoRI, and 25 ìl of de-ionized water. The mixture was incubated for two h at 37 ºC and placed at 70 ºC to inactivate the enzyme reaction. Once inactivation was complete 24 ìl of adaptors solution (Life Technologies Gibco BRL) was added to the digestion along with T4 DNA ligase (3 U/µl). The cocktail was incubated at 20 ºC for 12 h then diluted to 0.1 X with TE, and stored at -20 ºC.
Pre-amplification reactions were accomplished by adding 5 µl of 10X PCR buffer, 1 U of Taq DNA polymerase for a final volume of 51 ìl. The PCR reaction was performed in Perkin-Elmer thermocycler (model 9600), using 94 ºC for 30 min, 56 ºC for 60 s, and 72 ºC for 60 s, for 20 cycles.
The primers used were MseI (GATGAGTCCTGAGT AA) and EcoRI (GACTGCGTACCAATTCCAA). The primer combinations were: (1) EcoRI AT- MseI CG, (2) EcoRI AT MseI CA, (3) EcoRI AT MseI CC, (4) EcoRI AA MseI CA, (5) EcoRI AA MseI CC, (6) EcoRI AA MseI CT, (7) EcoRI AA MseI CG. The primers were labeled with 33P in 50 ìl reactions containing 0.1 ml of a 50 ng/ml of EcoRI primer + two base pairs, 10ìl of de-ionized water, 10 ìl of 5X kinase buffer, 10 ìl of ãP33 (3.000 Ci/mmol), 0.2 U of T4 kinase. The reagents were mixed and incubated for one hour at 37 ºC. At the end of the reaction time the mixture was inactivated at 70 ºC.
Amplification reactions were performed using 50 ng/ml of EcoRI and of MseI primer, 5 mM of dNTPs, for a final volume of 50 ìl. Added to another tube were 79 ìl of de-ionized water, 20 ìl of PCR buffer 10X, and DNA Taq polymerase (5 units/ìl ) for a total volume of 100 ìl. For amplifications, 5 µl of the pre-amplification reaction (diluted 1:50 with TE buffer) were used. The reactions were performed in a Perkin-Elmer thermocycler using 94 ºC for 30 s, 65 ºC for 30 s, and 72 ºC for 60 s. The thermocycler was programmed for 12 cycles with a 0.7 ºC decrease in the annealing temperature after each cycle, followed by 23 cycles of 94 ºC/30 s, 56 ºC/30 s and 72 ºC/60 s.
After amplification, 20 ìl of formamide mix (98% formamide, 10 mM EDTA, 1% bromophenol blue and 1% cianole xylene), were added to each reaction. DNA products were separated on 12.5% denaturing polyacrilamide gel electrophoresing at 75W for approximately 3 h then dried under vacuum on Whatman 3MM paper for 30 min, and exposed to X ray film for up to ten days.
Mycelia from C. gossypii were pink, compact with short dense setae; and short hyaline conidiophores. Mycelia from C. gossypii var. cephalosporioides were gray, aerial, with long sparse setae covered with conidial mass and long aerial conidiophores.
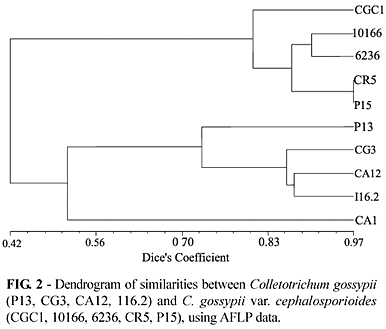
All isolates resulting in ramulose showed symptom scores ranging from 3 to 5, depending on the cotton cultivars. Isolates CGC1, 10166, 6236, CR5 and P15 were identified as C. gossypii var. cephalosporioides. Plant symptoms consisted of stunted internodes, twisted and enlarged branches, wrinkled deformed leaves with chlorotic areas that evolved to rounded elongated and angular necrotic lesions, sometimes accompanied by perforations. C. gossypii isolates P13, CG3, CA1, CA12, and I16.2 did not cause any of the symptoms described above.
The AFLP analyses showed a total of 318 polymorphic and 16 monomorphic bands, revealing clear distinction between the two pathogenic forms of the ten C. gossypii tested (Figure 1).
Morphological characterization and AFLP marker testing aimed at distinguishing C. gossypii from C. gossypii var. cephalosporioides. Even though morphological characteristics demonstrated differences between anthracnose and ramulose causal agents, AFLP markers showed an additional advantage by detecting genetic diversity within pathogen populations from small amounts of DNA without doing time-consuming pathogen culturing. Other studies have shown the discriminatory power of the AFLP technique for Colletotrichum species (O'Neill et al., 1997).
Anthracnose isolates were grouped at 51.2% similarity compared to 80.4% for the ramulose isolates (Figure 2). The greater similarity suggests a closer relationship among the ramulose isolates. It is possible that aggressive isolates may be required to withstand higher selection pressures in nature, with ramulose isolates being more agressive than anthracnose isolates. A second hypothesis would be that ramulose isolates evolved from anthracnose isolates thus having a narrower genetic base.
The ramulose isolates, CR5 and P15, obtained from two different plants in the same field, were found to have the closest similarity at 97%. The ramulose isolate 10166 obtained from cotton and 6263 from Picão Preto (Bidens pilosa L.) presenting 90.53% of similarity. Colletotrichum gossypii var. cephalosporioides can infect different host species, which in evolutionary terms is a favorable aspect for dissemination and adaptation to adverse environmental conditions. As it remains in crop residues and weeds, from an agronomic point of view, there are important implications for cotton cropping and disease control strategies. The anthracnose isolate "CA1" was the most divergent with an average similarity of 51.3%, when compared to the other anthracnose isolates. Similar results were obtained for the ramulose CGC1 isolate, which was the most divergent (average similarity of 81.1%) within the ramulose group. The dendrogram (Figure 2) shows the separation of the twopathogenic forms of C. gossypii in two groups at 42% similarity level. Our results clearly showed the discriminatory power of the AFLP technique in differentiating between the anthracnose and ramulose causal agents.
LITERATURE CITED
Accepted for publication on 08/07/2004
Corresponding author: Renata Silva-Mann
- BAILEY, J.A. & JEGER, M.J. Colletotrichum: biology, pathology and control. Wallingford. CAB International. 1992.
- CIA, E. Ocorrência e conhecimento das doenças do algodoeiro anual Gossypium hirsutum L. no Brasil. Summa Phytopathologica 3:167-193. 1977.
- DAVIS, R.G., BIRD, L.S., CHAMBERS, A.Y., GARBER, R.H., HOWELL, C.R., MINTON, E.B., STERNE, R. & JOHNSON, L.F. Seedling disease complex. In: Watkins, G.M. (Ed.) Compendium of cotton diseases. St. Paul. American Phytopathology Society, 1981. pp.1-87.
- DICE, L.R.. Measures of the amount of ecologic association between species. Ecology 26:297-302. 1945.
- MILLS, P.R., SREENIVASAPRASAD, S. & BROWN, A.E. Detection of the Anthracnose Pathogen Colletotrichum In: Schots, A., Dewey, F.M. & Oliver, R. (Eds.) Modern assays for plant pathogenic fungi: identification, detection and quantification. Wallingford. Redwood Press. 1994. pp.183-189.
- O'NEILL, N.R, VAN BERKUM, P.B., LIN, J.J., KUO, J., UDE, G.N., KENWORTHY, W. & SAUNDERS, J.A. Application of Amplified Fragment Length Polymorphism (AFLP) for the genetic characterization of Colletotrichum pathogens of alfalfa (Medicago sativa). Phytopathology 87:745-750. 1997.
- ROHLF, F.J. NTSYS-PC: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software: Setauket, New York. 2000.
- TANAKA, M.A.S. Problemas da detecção do agente causal da ramulose em sementes de algodão. In: Menten, J.O.M. (Ed.) I Patógenos em sementes: detecção, danos e controle químico. São Paulo. Ciba Agro. 1995. pp.93-108.
- TANAKA, M.A.S., MENTEN, J.O.M. & MACHADO, J.C. Hábito de crescimento de Colletotrichum gossypii e C. Gossypii var. cephalosporioides em sementes de algodoeiro. Bragantia 55:5-104. 1996.
- TEIXEIRA, H. Variabilidade de Acremonium strictum e sua trasmissibilidade e efeitos em sementes de milho (Zea mays L.). (Tese Doutorado). Lavras. Universidade Federal de Lavras. 2001.
- ZABEAU, M. Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application n. 0534858 A1. 1993.
Publication Dates
-
Publication in this collection
17 May 2005 -
Date of issue
Apr 2005
History
-
Accepted
08 July 2004