Abstract
The aim of this research was to study the mating system of a natural population of Theobroma grandiflorum (cupuassu) from Nova Ipixuna, Pará state, using microsatellite markers. Eight polymorphic microsatellite loci were analyzed in eight families, each represented by 10 six-month old seedlings derived from open-pollinated pods. The estimation for the multilocus outcrossing rate (<img border=0 id="_x0000_i1026" src="../../img/revistas/gmb/v26n3/a25img01.gif" align=absmiddle>m = 1.0) and individual outcrossing rate (<img border=0 id="_x0000_i1027" src="../../img/revistas/gmb/v26n3/a25img01.gif" align=absmiddle> = 1.0) for this population suggests that T. grandiflorum may be a perfect outbreeding (allogamous) species. Likewise, for the studied population the estimate for single locus outcrossing rate (<img border=0 id="_x0000_i1028" src="../../img/revistas/gmb/v26n3/a25img01.gif" align=absmiddle>S) was elevated (0.946), but lower than <img border=0 id="_x0000_i1029" src="../../img/revistas/gmb/v26n3/a25img01.gif" align=absmiddle>m, confirming the likely outcrossing character of the species and suggesting the occurrence of 5.4% biparental inbreeding rate (<img border=0 id="_x0000_i1030" src="../../img/revistas/gmb/v26n3/a25img01.gif" align=absmiddle>m - <img border=0 id="_x0000_i1031" src="../../img/revistas/gmb/v26n3/a25img01.gif" align=absmiddle>S). The estimation of genetic divergence (<img border=0 id="_x0000_i1032" src="../../img/revistas/gmb/v26n3/a25img02.gif" align=absmiddle>st) between allelic frequencies in ovules and pollen revealed a deviation from random mating in 75% of the evaluated loci. Likewise, the estimate of correlation of paternity (<img border=0 id="_x0000_i1033" src="../../img/revistas/gmb/v26n3/a25img03.gif" align=absmiddle>P = 0.930) and the mean coefficient of co-ancestrality within families (<img border=0 id="_x0000_i1034" src="../../img/revistas/gmb/v26n3/a25img04.gif" align=absmiddle>XY = 0.501) indicated that the outcrossings were predominantly correlated, and the offspring were full-sibs. These results suggested that for this particular population of T. grandiflorum, the sampling strategy for genetic conservation and breeding should adopt specific models for families derived from correlated outcrossing (full-sibs) and not the ones usually adopted in classic outcrossing species breeding programs (half-sibs).
Amazonia; germplasm; cupuassu; genetic conservation; simple sequence repeat
RESEARCH ARTICLE
Mating system in a natural population of Theobroma grandiflorum (Willd. ex Spreng.) Schum., by microsatellite markers
Rafael M. AlvesI, II; Angela S. ArteroIII; Alexandre M. SebbennIV; Antonio FigueiraIII
IEmbrapa Amazônia Oriental, Belém, PA, Brazil
IIEscola Superior de Agricultura "Luiz de Queiroz", Departamento de Genética, Piracicaba, SP, Brazil
IIIUniversidade de São Paulo, Centro de Energia Nuclear na Agricultura, Piracicaba, SP, Brazil
IVInstituto Florestal de São Paulo, São Paulo, SP, Brazil
Correspondence Correspondence to Antonio Figueira Universidade de São Paulo, Centro de Energia Nuclear na Agricultura Av. Centenário 303 Caixa Postal 96, 13400-970 Piracicaba, SP, Brazil E-mail: figueira@cena.usp.br
ABSTRACT
The aim of this research was to study the mating system of a natural population of Theobroma grandiflorum (cupuassu) from Nova Ipixuna, Pará state, using microsatellite markers. Eight polymorphic microsatellite loci were analyzed in eight families, each represented by 10 six-month old seedlings derived from open-pollinated pods. The estimation for the multilocus outcrossing rate ( m = 1.0) and individual outcrossing rate (
m = 1.0) and individual outcrossing rate ( = 1.0) for this population suggests that T. grandiflorum may be a perfect outbreeding (allogamous) species. Likewise, for the studied population the estimate for single locus outcrossing rate (
= 1.0) for this population suggests that T. grandiflorum may be a perfect outbreeding (allogamous) species. Likewise, for the studied population the estimate for single locus outcrossing rate ( S) was elevated (0.946), but lower than
S) was elevated (0.946), but lower than  m, confirming the likely outcrossing character of the species and suggesting the occurrence of 5.4% biparental inbreeding rate (
m, confirming the likely outcrossing character of the species and suggesting the occurrence of 5.4% biparental inbreeding rate ( m -
m -  S). The estimation of genetic divergence (
S). The estimation of genetic divergence (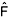 st) between allelic frequencies in ovules and pollen revealed a deviation from random mating in 75% of the evaluated loci. Likewise, the estimate of correlation of paternity (
st) between allelic frequencies in ovules and pollen revealed a deviation from random mating in 75% of the evaluated loci. Likewise, the estimate of correlation of paternity (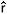 P = 0.930) and the mean coefficient of co-ancestrality within families (
P = 0.930) and the mean coefficient of co-ancestrality within families (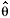 XY = 0.501) indicated that the outcrossings were predominantly correlated, and the offspring were full-sibs. These results suggested that for this particular population of T. grandiflorum, the sampling strategy for genetic conservation and breeding should adopt specific models for families derived from correlated outcrossing (full-sibs) and not the ones usually adopted in classic outcrossing species breeding programs (half-sibs).
XY = 0.501) indicated that the outcrossings were predominantly correlated, and the offspring were full-sibs. These results suggested that for this particular population of T. grandiflorum, the sampling strategy for genetic conservation and breeding should adopt specific models for families derived from correlated outcrossing (full-sibs) and not the ones usually adopted in classic outcrossing species breeding programs (half-sibs).
Key words: Amazonia, germplasm, cupuassu, genetic conservation, simple sequence repeat.
Introduction
The mating system of a plant species determines how the genetic information is transferred from one generation to the next (Wright, 1921), fundamental information for genetic conservation and breeding programs. The knowledge about how genes will be combined in the next generation allows strategies to be established for selection of superior genotypes and to keep genetic variability for future use. In tropical ecosystems, earlier studies mistakenly suggested that self-fertilization was the predominant mating system, considering the asynchronous flower opening among plants (Baker, 1959), or the difficulty of pollinator mobility through the complex structure of the dense forest (Fedorov, 1966). Only after the study on pollen flow over long distances by Jansen (1971) and surveys conducted by Bawa (1974), it was concluded that tropical trees present diverse mating strategies to avoid self-fertilization and favor outcrossing. In general, tropical trees are considered to possess a mixed mating system, mostly outbreeding (Murawski and Hamrick, 1992). Most flowers of tropical species are hermaphrodite (Bawa, 1974; Silva 1996), but there is a lack of information about the mating systems, especially for Amazonian species.
Theobroma grandiflorum (Willd. ex Spreng.) Schum. (cupuassu), is an important fruit tree, native to the Brazilian Amazon rainforest with its putative center of diversity located at the South/Southeastern region of Pará state (Cuatrecasas, 1964). Cupuassu is diploid (2n = 20), relative of cacao (T. cacao), the species of major economic importance of the genus Theobroma (Malvaceae, sensu latu). The strong tropical flavor from the seed-surrounding pulp is highly appreciated in juices, ice cream, jams, candies, desserts and liquors (Calzavara et al., 1984). The seed-pulp represents up to 40% of the fruit weight, while a product similar to cocoa powder (called "cupulate") can be obtained from fermented seeds (Ribeiro et al., 1992).
Even in areas of naturally high cupuassu incidence, the highest tree density is around 1 to 2 individuals ha-1, reaching 3.75 plants ha-1 (Homma et al., 2001). Under natural Amazonian conditions, cupuassu blooms during the dry season (July to December), and produces fruits from August to April, with a major peak from January to March, the local rainy season (Prance and Silva, 1975). Cupuassu flowers are hermaphrodite, but present extremely efficient anatomical barriers to avoid direct access of pollen grains from anthers to the stigma. Silva (1996) observed that 100% of spontaneous self-fertilization was avoided based only on hercogamy, and concluded that T. grandiflorum did not present agamospermy requiring pollination for pod set. In the related species Theobroma cacao (cacao), the presence of flower physical barriers favors cross-pollination, even for self-compatible individuals (Souza and Dias, 2001). All attempts to artificially self-fertilize cupuassu have failed (Silva, 1996; Alves et al., 1997a), indicating that T. grandiflorum should also present a self-incompatibility mechanism with a strong trend for outcrossing as its predominant mating system (Venturieri, 1993; Venturieri and Ribeiro Filho, 1995; Silva, 1996; Alves et al., 1997a). Addison and Tavares (1951) described a cupuassu tree, which was self-compatible and allo-compatible, able to cross-pollinate other Theobroma species.
Self-incompatibility is a common phenomenon in tropical species as an efficient system to avoid inbreeding (Bullock, 1985). Theobroma species are no exception, and self-incompatibility has been widely described for T. cacao (Cope, 1962; Knight and Rogers, 1955). Cacao presents a unique incompatibility system, in which the incompatibility reaction does not occur at the style and stigma or by inhibition of pollen tube growth, but by failure of gamete nuclei fusion in the embryo sac. This system exhibits sporophytic and gametophytic features. Incompatible matings, which lead to flower abscission 72 h after pollination, are characterized by the presence of non-fused nuclei in the ovary. Conversely, Aneja et al. (1994) worked with a self-compatible cacao genotype (IMC 30) and proposed that the self-incompatibility system occurs in two stages: pollen germination and gametic fusion. Similarly to Theobroma cacao, T. grandiflorum might present a pre-zygotic self-incompatibility system leading to flower abscission before zygote formation (Venturieri, 1994).
Studies on mating systems have benefited from the development of biochemical markers, such as isozymes, since the 1970s. However, with the development of DNA markers, the establishment of mating systems became more precise. Microsatellite markers present great advantages for determining mating systems since these are co-dominant markers, multi-allele, robust, reliable and simple (Ferreira and Grattapaglia, 1996). Allele frequencies can be estimated directly without requiring genetic crosses (Hamrick et al., 1979).
The objective of this work was to define the natural mating system of Theobroma grandiflorum based on allele segregation of microsatellite loci in families derived from natural open-pollinated crosses in a wild population from Nova Ipixuna, Pará state, at the species putative center of diversity.
Material and Methods
Plant material
Samples were collected from a naturally occurring T. grandiflorum population from a dense rainforest in Nova Ipixuna, Pará state (4°53' S; 49°22' W). Budwood and leaves were collected from 40 mature trees in December 2001, and a random sub-sample of eight trees (bearing pods at the time of collection), together with ten 6-month-old seedlings from each individual were used in the present analysis.
DNA extraction
Samples of ca. 150 mg of leaf tissue were ground in liquid nitrogen and DNA was extracted using a protocol adapted from Doyle and Doyle (1990), described by Figueira et al. (1997). Briefly, ground leaf tissues were extracted with buffer [2% CTAB; 1.4 M NaCl; 100 mM Tris-HCl, pH 8.0; 20 mM EDTA pH 8.0; 1% polyvinylpyrrolidone MW10, 000; 0.2% b-mercaptoethanol; and 0.1 mg.mL-1 of proteinase K], mixed well and incubated at 55 °C for 60 min. The solution was then extracted three times with chloroform: isoamyl-alcohol (24:1 v/v). DNA was precipitated from the aqueous phase adding an equal volume of cold isopropanol. The DNA was centrifuged, washed with 70% Ethanol to remove salts and allowed to air dry. The DNA pellet was resuspended in 20 mL TE buffer (10 mM Tris-HCl, pH 8; 0,1 mM EDTA) containing ribonuclease A (10 mg.mL-1) and incubated at 37 °C for 30 min. DNA was quantified by fluorimetry in a DyNA Quant 2000 fluorometer (Amersham Biosciences, Buckinghamshire, UK) and a 5 ng mL-1 stock was prepared.
Microsatellite analyses
The total reaction volume was 13 mL, containing 15 ng of genomic DNA; 50 mM KCl; 10 mM Tris-HCl (pH 8.8); 0.1% Triton X-100; 1.5 mM MgCl2; 100 mM of each dNTPs; 0.2 mM of each primer and 1.2 units of Taq polymerase (Invitrogen do Brasil, São Paulo, Brazil). Ten primer-pairs (referred as primers throughout the text) developed for T. cacao by Lanaud et al. (1999) were used. The primers tested were synthesized by Invitrogen do Brasil. Amplifications were conducted on the Perkin Elmer GeneAmp 9600 or 9700 thermocycler (Applied Biosystems, Foster City, CA, USA), programmed initially as Lanaud et al. (1999) with a denaturing step at 94 °C for 4 min, followed by 30 cycles of 40 s at 94 °C; 60 s at 46 or 51 °C; and 60 s at 72 °C. Sequencing gels (6% polyacrylamide; 7 M urea) were run in TBE under standard conditions at 55 W for 2 h and the products were visualized by silver staining according to the procedure described by Creste et al. (2001).
Statistical analyses
The mating system was analyzed based on the mixed mating model (Ritland and Jain, 1981) and sibling-pair model (Ritland, 1989) using the software MLTR (Ritland, 1997). The estimated parameters were: 1. Multilocus outcrossing rate (
m) by maximum-likelihood using the expectation-maximization method; 2. Single locus outcrossing rate (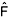 s); 3. Biparental inbreeding (
s); 3. Biparental inbreeding (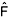 m -
m - 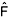 s); 4. Individual multilocus outcrossing rate (
s); 4. Individual multilocus outcrossing rate (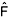 ); 5. Pollen and ovules allele frequencies; 6. Fixation index for maternal genotypes (
); 5. Pollen and ovules allele frequencies; 6. Fixation index for maternal genotypes (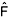 m); 7. Correlation of outcross paternity (1/
m); 7. Correlation of outcross paternity (1/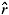 p); and 8. Probable number of pollinators (1/
p); and 8. Probable number of pollinators (1/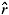 p). The assumptions of the mixed mating model were the same as reported by Ritland and Jain (1981). The standard error of the estimated parameters was obtained by 1,000 re-sampling bootstraps.
p). The assumptions of the mixed mating model were the same as reported by Ritland and Jain (1981). The standard error of the estimated parameters was obtained by 1,000 re-sampling bootstraps.
The test for random outcrossing was initially conducted by testing allelic frequencies of pollen and ovules for heterogeneity by estimating
ST (Wright, 1965), used as a measure of genetic divergence between allele frequencies of the distinct groups. A chi-square test was employed to test the significance for each locus [c2 = 2nST(k-1); DF = (k-1)(s-1)], proposed by Workman and Niswander (1970), where: n = number of gametes in both groups (pollen and ovules), k = number of alleles; and s = number of groups (in our case 2, pollen and ovules). The fixation index for all families (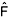 F) and the mean co-ancestrality coefficient among plants within families (
F) and the mean co-ancestrality coefficient among plants within families (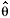 xy) were obtained from the analysis of variance of gene frequencies, and the confidence intervals were estimated by 10,000 re-sampling bootstraps, estimated using the software GDA (Lewis and Zaykin, 1999).
xy) were obtained from the analysis of variance of gene frequencies, and the confidence intervals were estimated by 10,000 re-sampling bootstraps, estimated using the software GDA (Lewis and Zaykin, 1999).
Results
All individuals from all families displayed at least one maternal allele, indicating that there was no seed contamination (data not shown). From the 10 microsatellite loci tested, eight were polymorphic, and two (CIR18 and CIR58) were monomorphic. The polymorphic loci segregated from 2 to 5 alleles, with a total of 25 alleles in the population (Table 1). No exclusive allele was detected either for pollen or for ovules from the crosses. However, the estimation of genetic divergence between allele frequencies from ovules and pollen were not homogeneous for 75% of the loci, revealing a deviation from random mating (Table 1). Heterogeneity of pollen allele frequencies among maternal genotypes was detected for 75% of the loci, indicating non-random sampling of the pollen pool by each maternal tree.
The individual multilocus outcrossing rate for each maternal genotype was equal to 1 and did not vary among trees (Table 2). The population multilocus outcrossing rate ( m) was equal to 1.0, indicating that T. grandiflorum appears to be a perfect outbreeding species. Similarly, the single locus outcrossing rate (
m) was equal to 1.0, indicating that T. grandiflorum appears to be a perfect outbreeding species. Similarly, the single locus outcrossing rate ( s) was high (0.946; Table 2), but significantly different from the multilocus outcrossing rate (1.0), considering the standard error of the mean (0.029), thus, suggesting that crosses among related individuals have occurred in the population. The estimated biparental inbreeding rate (
s) was high (0.946; Table 2), but significantly different from the multilocus outcrossing rate (1.0), considering the standard error of the mean (0.029), thus, suggesting that crosses among related individuals have occurred in the population. The estimated biparental inbreeding rate ( m -
m -  s) was low (5.4%), but significantly different from zero, considering the standard error of the mean (Table 2).
s) was low (5.4%), but significantly different from zero, considering the standard error of the mean (Table 2).
The fixation index for maternal genotypes (
m = 0.008) and within progenies (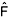 p = -0.096) were low and not significant, indicating the absence of inbreeding (Table 2). The mean co-ancestrality coefficient among plants within families was close to the expected values from full-sib families (
p = -0.096) were low and not significant, indicating the absence of inbreeding (Table 2). The mean co-ancestrality coefficient among plants within families was close to the expected values from full-sib families (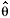 xy = 0.251), and it was significantly different from zero, based on standard error. This result supports the hypothesis of bias from random mating. Similarly, the correlation of paternity (
xy = 0.251), and it was significantly different from zero, based on standard error. This result supports the hypothesis of bias from random mating. Similarly, the correlation of paternity (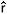 p) was high and significantly different from zero (0.930), indicating that many of the offspring were derived from related crosses, sharing the same paternal and maternal genitor. The estimation of the probable number of pollinators (1/
p) was high and significantly different from zero (0.930), indicating that many of the offspring were derived from related crosses, sharing the same paternal and maternal genitor. The estimation of the probable number of pollinators (1/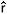 p = 1,07) was slightly superior to one, suggesting that, on average, two pollinators contributed to the crosses.
p = 1,07) was slightly superior to one, suggesting that, on average, two pollinators contributed to the crosses.
Discussion
The estimation of the multilocus outcrossing rate for the Nova Ipixuna natural population suggested that T. grandiflorum may be a perfect outbreeding species (1.0). Similarly, the estimation of the individual multilocus outcrossing rate was equal to one for all the maternal genotypes. The outcrossing rate for hermaphroditic species depends on the presence and intensity of self-incompatibility mechanisms; the degree of protoandry or protogyny; pollinators foraging behavior between and within the trees, which is affected by flowering density in the population and selective abortion of selfed seeds or fruits (Murawski and Hamrick, 1991). Previous studies suggested that T. grandiflorum was probably an outbreeder species (Venturieri, 1994; Alves et al., 1996; Silva, 1996). The T. grandiflorum flower anatomic structure presents physical barriers isolating the stigma from anthers, in addition to the complex self-incompatibility system, indicating that the species should be an outbreeder (Venturieri, 1993; Venturieri and Ribeiro, 1995). Self-incompatibility systems seem to be common to all Theobroma species, and have been particularly studied in T. cacao (Cope, 1962; Knight and Rogers, 1955; Yamada et al., 1982). Similarly to cacao, T. grandiflorum might present the self-incompatibility reaction at the ovary, pre-zygotic, leading to flower abscission before embryo formation. The absence of selfing in T. grandiflorum has been reported by various authors using either flower bud isolation (spontaneous selfing) or by controlled pollination with pollen from the same tree (Falcão and Lleras, 1983; Machado and Retto Júnior, 1991; Alves et al., 1996; Silva, 1996). All results indicated that cupuassu presents a self-incompatibility system, but the elucidation of possible genetic control of this mechanism requires further studies.
The high multilocus outcrossing rate estimated for this natural population of T. grandiflorum (1.0) is comparable to ones described for other tropical tree species (Murawski et al., 1990; Murawski and Hamrick, 1992; Sebbenn et al., 2000; Seoane et al. 2001). According to Seoane et al. (2001), the occurrence of selfing in predominantly outbreeder species derives from deficiencies in the mechanism of self-incompatibility during the flowering period, favoring selfing during specific phases. For T. grandiflorum, Silva (1996) did not observe any variation in self-incompatibility during the flowering period. In general, tropical tree species have demonstrated that they have a mixed mating system with the predominance of outbreeding and an outcrossing rate above 85%. Plants with long life span, such as these trees, tend to accumulate large somatic mutations and selfing could lead to extinction. Thus Lande and Schemske (1985) hypothesized that natural selection might have driven these species to a high outcrossing rate.
The estimation of genetic divergence (
ST) between the allele frequencies of pollen and ovules (maternal x paternal) indicated deviation from random mating for most of the loci (75%). Differences in allele frequencies between maternal and paternal genotypes have been observed in various studies on tree species mating systems (Murawski et al., 1990; Khasa et al., 1993; Lee et al., 2000; Sebbenn et al., 2000; Rocha and Aguilar, 2001). These deviations in a perfect outbreeder species (t = 1) have important implications on breeding and conservation strategies. The deviations increase the levels of within-progeny relationship above that expected from random outcrossing, overestimating genetic parameters, such as heritability and genetic gains for selection (Namkoong, 1966; Squillace, 1974). In genetic conservation programs, these deviations imply that sample size should be larger than retained if crosses were random, since the increased relationship within families reduces the effective population size.The correlation of the paternity estimate was high (rp = 0.930), suggesting that most of the offspring were full-sibs. Few studies with tropical tree species have reported estimation of correlation of paternity. The rp estimation for Esenbeckia leiocarpa populations ranged from 0.749 to 0.986 (Seoane et al., 2001). For Cariniana legalis populations, the correlation of paternity estimated ranged from 0.212 to 0.324, while for Tabebuia cassinoides, it varied from 0.295 to 0.547 (Sebbenn et al., 2001). The causes for correlated outcrossing might derive from pollinator behavior, visiting mostly neighboring trees; polyandry or deposit of multiple pollen grains from one pollinator; small number of close neighboring trees (Sun and Ritland 1998); or flower asynchrony among trees (Murawski, 1995). Many of these factors might have contributed to the high rate of correlated outcrossing observed for T. grandiflorum. Considering that pollen and stigma viability are practically the same in cupuassu (Silva, 1996), the low population density, associated with flowering asynchrony, might be the likely cause for the observed correlated outcrossing. Alves et al. (1997b) investigated an ex situ collection of T. grandiflorum and detected significant differences among accessions for flower production and blossom period. Murawski and Hamrick (1991) observed that for low-density populations of trees, the pollen in crosses were less diversified (multiple origins) than in higher density populations. Environmental variations can affect pollinator behavior and/or density, affecting the outcrossing rate of the species in distinct populations (Murawski, 1995).
The estimated multilocus outcrossing rate (1.0), combined with the high paternity correlation (rp) allowed to conclude that approximately 93% [ m rp] of the offspring derived from correlated outcrossing (full-sibs) and only 7% [
m rp] of the offspring derived from correlated outcrossing (full-sibs) and only 7% [ m (1 - rp)] derived from random mating, related as half-sibs. The estimated number of effective pollinators (1/rp) did not exceed to two. Thus, the T. grandiflorum families in this population are mainly full-sibs. Likewise, the estimated value for co-ancestrally within families (
m (1 - rp)] derived from random mating, related as half-sibs. The estimated number of effective pollinators (1/rp) did not exceed to two. Thus, the T. grandiflorum families in this population are mainly full-sibs. Likewise, the estimated value for co-ancestrally within families (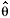 xy = 0.251) was close to the expected value for full sib families (0.25).
xy = 0.251) was close to the expected value for full sib families (0.25).
No inbreeding was detected for the maternal genotypes (
m = 0.008) and families (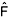 F = -0.096). Because it was estimated that 5.4% of crosses occurred among relatives, a small inbreeding rate was expected.
F = -0.096). Because it was estimated that 5.4% of crosses occurred among relatives, a small inbreeding rate was expected.
Families from open-pollinated crosses are mostly considered as random mating, resulting in half-sibs. The observed results discard this hypothesis, demonstrating that most families are full-sibs. As a result, the additive genetic variance and related parameters, such as heritability and selection gain will be overestimated if the families are assumed to be half-sibs (Namkoong, 1966; Squillace, 1974). In families derived from open-pollination, collected from populations without inbreeding and relationships, the genetic correlation within families (rxy) estimates one fourth of the additive genetic variance (Namkoong 1966). However, if correlated outcrossing occurred, such as observed here, the correlation rxy, is not one fourth. Substituting the estimated parameters in Table 2, rxy in the model proposed by Ritland (1989) for species with mixed mating system  , the estimated value is equal to 0.482, about twice the value expected for half-sib families (0.25). Further, the correlation coefficient is twice the co-ancestrality coefficient among plants within families (Falconer and Mackay, 1997), resulting in
, the estimated value is equal to 0.482, about twice the value expected for half-sib families (0.25). Further, the correlation coefficient is twice the co-ancestrality coefficient among plants within families (Falconer and Mackay, 1997), resulting in 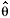 xy = 0.243, similar to the coefficient of co-ancestrality estimated by the analyses of variance of allele frequencies (0.251; Table 2), confirming the conclusion that the families are full-sibs.
xy = 0.243, similar to the coefficient of co-ancestrality estimated by the analyses of variance of allele frequencies (0.251; Table 2), confirming the conclusion that the families are full-sibs.
The mixed system strongly affects the estimation of effective population size (Ne). For instance, assuming absence of inbreeding and relationships in the parental generation, and collecting an infinite number of seeds from eight trees, the effective size would be approximate to 4m, where m is the number of families, and  e = 32. On the other hand, based on the same previous conditions, using the equation of effective size of variance derived from Cockerham (1969) for hermaphrodite species with mixed system, then
e = 32. On the other hand, based on the same previous conditions, using the equation of effective size of variance derived from Cockerham (1969) for hermaphrodite species with mixed system, then e = 0,5 / [
e = 0,5 / [ 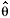 xy ( n 1 ) / n + (1 +
xy ( n 1 ) / n + (1 + 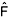 p) / 2n] where
p) / 2n] where 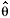 xy is the average co-ancestrality coefficient within the family; n is the total sample size (assuming infinite 1.000), and
xy is the average co-ancestrality coefficient within the family; n is the total sample size (assuming infinite 1.000), and 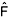 p is the fixation index in the families. Substituting the values estimated in Table 2, the effective size is 16.5. Thus, the estimated effective population size considering the parameters of the mating system is about 50% less than expected from random mating (
p is the fixation index in the families. Substituting the values estimated in Table 2, the effective size is 16.5. Thus, the estimated effective population size considering the parameters of the mating system is about 50% less than expected from random mating ( e = 32). These results demonstrate that it is fundamental to have previous knowledge of mating systems of a species in breeding programs and ex situ genetic conservation programs. Therefore, for T. grandiflorum genetic conservation, it would be necessary to collect seeds of about 25 trees to retain the effective size of 50, a normal standard used for short term conservation.
e = 32). These results demonstrate that it is fundamental to have previous knowledge of mating systems of a species in breeding programs and ex situ genetic conservation programs. Therefore, for T. grandiflorum genetic conservation, it would be necessary to collect seeds of about 25 trees to retain the effective size of 50, a normal standard used for short term conservation.
In conclusion, our results based in this specific natural population, suggested that T. grandiflorum may be a predominantly outbreeding species and the data suggested that the outcrossings were predominantly correlated, with full-sib offspring. These results should be considered to define the sampling strategy for T. grandiflorum. genetic conservation and breeding. Definite conclusion about the T. grandiflorum mating system requires further analyses of other natural populations.
Acknowledgments
This research was supported by Embrapa Amazônia Oriental; International Foundation for Science (D2814-2) and CNPq (479573/2001-8). The authors thank the Brazilian funding agencies CNPq (R.M.A.; A.F.) and FAPESP (A.S.A.) for fellowships.
Editor: Natal Antonio Vello
Received: December 9, 2002
Accepted: June 12, 2003
- Addison GO and Tavares RM (1951) Observações sobre as espécies do gênero Theobroma que ocorrem na Amazônia. Boletim Técnico 25, Belém, Instituto Agronômico do Norte, 18 p.
- Alves RM, Oliveira RP de, Lima RR, Neves MP, Chaves JP, Araújo, DG de and Pimentel L (1996) Pesquisas com recursos genéticos e melhoramento do cupuaçuzeiro, em desenvolvimento na EMBRAPA/CPATU. 1 Workshop sobre as culturas da pupunha e cupuaçu, Manaus, Brazil. EMBRAPA, CPAA, p 127-135.
- Alves RM, Araújo DG and Fernandes JRQ (1997a) Compatibilidade entre clones de cupuaçuzeiro (Theobroma grandiflorum). Revista Brasileira de Genética 20:148.
- Alves RM, Corrêa JRV and Rodrigo M (1997b) Melhoramento genético do cupuaçuzeiro (Theobroma grandiflorum) no Estado do Pará. 1 Seminário Internacional sobre pimenta-do-reino e cupuaçu, Belém, Brazil. EMBRAPA, CPATU; JICA, 1997a. p 127-146. (Documentos, 88).
- Aneja M, Gianfagna T, Ng E and Badilla I (1994) Carbon-dioxide treatment partially overcomes self-incompatibility in a cacao genotype. Hortscience 29:15-17.
- Baker HG (1959) Reproductive methods as factors in speciation inflowering plants. Cold Spring Harbor Symposia on Quantitative Biology 24:177-191.
- Bawa KS (1974) Breeding systems of tree species of a lowland tropical community. Evolution 28:85-92.
- Bullock SH (1985) Breeding systems in the flora of tropical deciduous forest in Mexico. Biotropica 17:287-301.
- Calzavara BBG, Muller CH and Kahwage ONC (1984) Fruticultura tropical: o cupuaçuzeiro - cultivo, beneficiamento e utilização do fruto. Belém, Brazil. EMBRAPA-CPATU, 101 p.
- Cope FW (1962) The mechanism of pollen incompatibility in Theobroma cacao L. Heredity 17:157-182.
- Creste S, Tulmann Neto A and Figueira A (2001) Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Molecular Biology Reporter 19:299-306.
- Cockerham, CC (1969) Variance of gene frequencies. Evolution 23:72-84.
- Cuatrecasas JA (1964) Cocoa and its aliies: a taxonomic revision of the genus Theobroma. Contribution from the United States National Herbarium, Washington 35(6)p 32-46.
- Doyle JJ and Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13-15.
- Falcão MA and Lleras E (1983) Aspectos fenológicos, ecológicos e de produtividade do cupuaçu (Theobroma grandiflorum Willd ex Spreng Schum). Acta Amazônica 13:725-735.
- Falconer DS and Mackay TFC (1997) Introduction to quantitative genetics. London, Longman, 463 p.
- Fedorov AA (1966) The structure of the tropical rain forest and speciation in the humid tropics. Journal of Ecology 54:1-11.
- Ferreira ME and Grattapaglia D (1996) Introdução ao uso de marcadores moleculares em análise genética. 2nd ed. EMBRAPA, CENARGEN, Brasília, 220 p.
- Figueira A, Lambert SV, Carpenter D, Pires JL, Cascardo JCM and Romaczyk L (1997) The similarity of cocoa flavour of fermented seeds from fingerprinted genotypes of Theobroma cacao from Brazil and Malaysia. Tropical Agriculture 74(2):132-139.
- Hamrick JL, Linhart YB and Mitton JB (1979) Relationships between life history characteristics and eletrophoretically detectable genetic variation in plants. Annual Review of Ecology and Systematic 10:173-200.
- Homma AKO, Carvalho RA and Menezes AJEA (2001) Extrativismo e plantio racional de cupuaçuzeiros no Sudeste Paraense: a transição inevitável. (Compact disc). 39 Congresso Brasileiro de Economia E Sociologia Rural, Recife, Brazil. Anais. SOBER, Brasília.
- Jansen DH (1971) Euglossine bees as long distance pollinators of tropical plants. Science 171:203-205.
- Khasa PD, Cheliak WM and Bousquet J (1993) Mating system of Racosperma auriculiforme in a seed production area in Zaire. Canadian Journal of Botany 71:779-785.
- Knight R and Rogers HH (1955) Incompatibility in Theobroma cacao Heredity 9:69-77.
- Lanaud C, Risterucci AM, Pieretti I, Falque M, Bouet A and Lagoda PJL (1999) Isolation and characterization of microsattelites in Theobroma cacao L. Molecular-Ecology 8:2141-2143.
- Lande R and Schemske DW (1985) The evolution of self-fertilization and inbreeding depression in plants. II Empirical observations. Evolution 39(1):41-52.
- Lee SL, Wickneswari R, Mahani MC and Zakri AH (2000) Mating system parameters in a tropical tree species, Shorea leprosula Miq. (Dipterocarpaceae), from Malaysian Lowland Dipterocarp Forest. Biotropica 32:693-702.
- Lewis PO and Zaykin D (1999) Genetic date analysis: computer program for the analysis of allelic date. Versão 1.0 para Windows 3.1. http//alleyn.eeb.uconn.edu/gda/
- Machado GME and Retto Jr AS (1991) Estudo preliminar sobre biologia floral do cupuaçuzeiro (Theobroma grandiflorum (Willd ex Spreng) Schum). Revista da Universidade do Amazonas. Série Ciências Agrárias 1:11-14.
- Murawski DA (1995) Reproductive biology and genetics of tropical trees from a canopy perspective. In: Lowman MD and Nadkarmi NM (eds) Forest Canopies. Academic Press, New York, pp 457-493.
- Murawski DA and Hamrick JL (1991) The effect of the density of flowering individuals on the mating systems of nine tropical tree species. Heredity 67:167-174.
- Murawski DA and Hamrick JL (1992) Mating system and phenology of Ceiba pentandra (Bombacaceae) in central Panama. Journal of Heredity 83:401-404.
- Murawski DA, Murawski JL, Hubbel SP and Foster RB (1990) Mating system of two Bombacaceous trees of a neotropical moist forest. Oecologia 82:501-506.
- Namkoong G (1966) Inbreed effects on estimation of genetic additive variance. Forest Science 12:8-13.
- Prance GT and Silva MF (1975) Árvores de Manaus. Manaus: INPA, 312 p.
- Ribeiro NCA, Sacramento CK, Barreto WS and Santos Filho LP (1992) Características físicas e químicas de frutos de cupuaçuzeiro (Theobroma grandiflorum) do sudeste da Bahia. Agrotrópica 4(2):33-37.
- Ritland K (1989) Correlated matings in the partial selfer Mimulus guttatus Evolution 43:848-859.
- Ritland K (1997) Multilocus mating system program MLTR. Version 1.1. University of British Columbia, Canadá. http//genetics.forestry.ubc.ca/ritland/programs.html
- Ritland K and Jain S (1981) A model for the estimation of outcrossing rate and gene frequencies using independent loci. Heredity 47:35-52.
- Rocha OJR and Aguilar G (2001) Variation in the breeding behavior of the dry forest tree Enterolobium cyclocarpum (Guanacaste) in Costa Rica. American Journal of Botany 89:1600-1606.
- Sebbenn AM, Kageyama PY, Siqueira ACMF and Zanatto ACE (2000) Taxa de cruzamento em populações de Cariniana legalis (Mart.) O. Ktze: Implicações para a conservação e o melhoramento genético. Scientia Forestalis 58:25-40.
- Sebbenn AM, Seoane CEC, Kageyama PY and Lacerta CM (2001) Estrutura genética em populações de Tabebuia cassiroides: Implicações para o manejo florestal e conservação genética. Revista do Instituto Florestal 13:99-111.
- Seoane CEC, Sebbenn AM and Kageyama PY (2001) Sistema reprodutivo em populações de Esenbeckia leiocarpa Revista do Instituto Florestal 13:21-28.
- Silva RM (1996) Estudo do sistema reprodutivo e divergência genética em cupuaçuzeiro (Theobroma grandiflorum Willd. ex Spreng.) Schum. Dissertação (Mestrado), Universidade de São Paulo, Escola Superior de Agricultura "Luiz de Queiroz", Piracicaba, 151 p.
- Souza CAS and Dias LAS (2001) Melhoramento ambiental e sócio-economia. Dias LAS (ed) Melhoramento genético do cacaueiro. FUNAPE, Viçosa, pp 1-48.
- Squillace AE (1974) Average genetic correlations among offspring from open-pollinated forest trees. Silvae Genetica 23:149-156.
- Sun M and Ritland K (1998) Mating system of yellow starthistle (Centaurea solstitialis), a successful colonizer in North America. Heredity 80:225-232.
- Venturieri GA (1993) Cupuaçu: A espécie, sua cultura, usos e processamento. Belém: Clube do cupu. 108 p.
- Venturieri GA (1994) Floral biology of cupuassu (Theobroma grandiflorum (Willdenow ex Sprengel) Schumann). PhD Thesis, University of Reading, Reading, 206 p.
- Venturieri GA and Ribeiro Filho AA (1995) Polinização manual do cupuaçuzeiro (Theobroma grandiflorum). Acta Amazônica 25:181-191.
- Yamada MM, Bartley BGD and Melo GRP (1982) Herança do fator compatibilidade em Theobroma cacao L. I. Relações fenotípicas na família PA (Parimari). Revista Theobroma 12:163-167.
- Workman P and Niswander JL (1970) Population studies on southwestern Indian tribes. II. Local genetic differentiation in the Papago. American Journal of Human Genetic 22:24-49.
- Wright S (1921) Systems of mating. Genetics 6:111-178.
- Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395-420.
Publication Dates
-
Publication in this collection
29 Sept 2003 -
Date of issue
2003
History
-
Accepted
12 June 2003 -
Received
09 Dec 2002



