Abstract
Gestation length, birth weight, and weaning weight of F2 Nelore-Angus calves (n = 737) with designed extensive full-sibling and half-sibling relatedness were evaluated for association with 34,957 SNP markers. In analyses of birth weight, random relatedness was modeled three ways: 1) none, 2) random animal, pedigree-based relationship matrix, or 3) random animal, genomic relationship matrix. Detected birth weight-SNP associations were 1,200, 735, and 31 for those parameterizations respectively; each additional model refinement removed associations that apparently were a result of the built-in stratification by relatedness. Subsequent analyses of gestation length and weaning weight modeled genomic relatedness; there were 40 and 26 trait-marker associations detected for those traits, respectively. Birth weight associations were on BTA14 except for a single marker on BTA5. Gestation length associations included 37 SNP on BTA21, 2 on BTA27 and one on BTA3. Weaning weight associations were on BTA14 except for a single marker on BTA10. Twenty-one SNP markers on BTA14 were detected in both birth and weaning weight analyses.
birth weight; gestation length; weaning weight; Nelore; whole genome association
ANIMAL GENETICS
RESEARCH ARTICLE
Alternative parameterizations of relatedness in whole genome association analysis of pre-weaning traits of Nelore-Angus calves
David G. Riley; Clare A. Gill; Andy D. Herring; Penny K. Riggs; Jason E. Sawyer; James O. Sanders
Department of Animal Science, Texas A&M University, College Station, TX, USA
ABSTRACT
Gestation length, birth weight, and weaning weight of F2 Nelore-Angus calves (n = 737) with designed extensive full-sibling and half-sibling relatedness were evaluated for association with 34,957 SNP markers. In analyses of birth weight, random relatedness was modeled three ways: 1) none, 2) random animal, pedigree-based relationship matrix, or 3) random animal, genomic relationship matrix. Detected birth weight-SNP associations were 1,200, 735, and 31 for those parameterizations respectively; each additional model refinement removed associations that apparently were a result of the built-in stratification by relatedness. Subsequent analyses of gestation length and weaning weight modeled genomic relatedness; there were 40 and 26 trait-marker associations detected for those traits, respectively. Birth weight associations were on BTA14 except for a single marker on BTA5. Gestation length associations included 37 SNP on BTA21, 2 on BTA27 and one on BTA3. Weaning weight associations were on BTA14 except for a single marker on BTA10. Twenty-one SNP markers on BTA14 were detected in both birth and weaning weight analyses.
Keywords: birth weight, gestation length, weaning weight, Nelore, whole genome association.
Introduction
Relatedness within animal data may be modeled in multiple ways ranging from incorporation of some aspect of family as a classification variable to more complex modeling of pedigree accomplished through inclusion of the numerator relationship matrix in mixed model equations (Henderson, 1963). The availability of dense single nucleotide polymorphism arrays for research permits the evaluation of genomic relatedness among animals with records based on their marker genotypes. Modeling of genomic relatedness, rather than average relatedness based on pedigree, was considered an important opportunity for genetic prediction (Liu et al., 2002; Visscher et al., 2006; Visscher, 2009), due to the improvement of regression equations through refinement of the covariances in the coefficient matrix that are due to relatedness among animals. Inclusion of genomic relatedness in prediction equations improved estimates of variances, and hence heritability (Thomas, 2005; Visscher et al., 2006; Wolc et al., 2013) and improved accuracies of predicted genetic merit (Hayes and Goddard, 2008). These appear to hinge upon the improved marker-trait associations detected with models using refined relationship models. Genomic relationship matrices incorporated in mixed model association analyses may be a better substitute for pedigree-based relationship in marker-QTL association studies (Yu et al., 2006; Zhang et al., 2006, 2007; Sham et al., 2009). There has not been comparison of association results using genomic relatedness and pedigree-based relatedness. The objectives of this study were to 1) describe the whole genome SNP-birth weight associations and the consequences of modeling relationship matrices derived either from pedigree or genomic information or modeling family structure as a fixed effect (per design of the experimental population), and 2) to describe weaning weight- and gestation length-SNP associations using records and genotypes of F2 Nelore-Angus calves.
Materials and Methods
Cattle
Calves (n = 737) were produced by embryo transfer at the Texas AgriLife Research Center near McGregor, Texas, USA. These calves were part of 14 Nelore-Angus full-sibling F2 families produced by five F1 Nelore-Angus sires (all F1 parents were Nelore-sired) and 10 F1 Nelore-Angus cows. The five sires were represented by 1, 2, 3, 4, and 4 families, respectively. The average number of calves in each family was 33.8 and ranged from 2 to 69. Calves were spring- and fall-born from the fall of 2003 through spring of 2007. Recipient cows consisted of 229 crossbred Bos indicus-Bos taurus cows; most had one calf, but a few had as many as four project calves. Four of those five F1 bulls were mated by natural service to 1/2 Bos indicus 1/2 Bos taurus cows to produce 4 half-sibling families. Most of the dams of calves in those half-sibling families were F1 or F2 Brahman-Hereford; some were Brahman-Angus and Nelore-Angus. All of those calves (n = 264) were spring-born (February to late April) from 2003 through 2007. Calves were identified and weighed as soon as practical after birth, usually within one day. Calves were weaned at an average of 213.8 ± 17.9 d of age. All procedures involving animals were approved by the Texas A&M University Institutional Animal Care and Use Committee with multiple animal use protocols from 2002 through the present.
Genotypes
Isolation of DNA was conducted from blood samples as previously described (Riley et al., 2013). Genotypes for all project animals were obtained from the Infinium BovineSNP50v1 assay (Illumina, Inc., San Diego, CA). There were on average 29,692 informative markers per F2 family (1 informative marker per 101 kb). Loci were excluded from association analyses if 1) genotypes were missing in 10% or more individuals because phasing programs and other software generally perform poorly with higher levels of missing genotypes, 2) minor allele frequency was less than or equal to 0.05 because very low minor allele frequencies could be SNP that were segregating in founders but not transmitted to the F1 and F2 individuals in the context of this experimental design (family structure), or 3) deviation from Hardy-Weinberg equilibrium (p < 0.05, exact tests according to Wigginton et al., 2005). The assay used is known to have approximately a 0.5% error rate (Hong et al., 2012); 684 Mendelian errors were detected in 181 of the 776 individuals (from 1 to 69 SNP in individuals).
Association analyses included 34,957 markers.
Statistical analyses
Models were built using the MIXED procedures of SAS (SAS Inst., Inc., Cary, NC). Investigated fixed effects included the main and interaction effects of year, season of birth, family, dam age (distinct categories for 2-, 3-, 4-, 5- to 10-yr-olds, and older than 10 yr), and calf gender. Regression of weaning weight on calf age in days was investigated. Association analyses were conducted using the Q-K procedures (Yu et al., 2006) of JMP Genomics (SAS Inst., Inc., Cary, NC) with fixed effects identified from the preliminary analyses and regression of trait on genotypic values, which were coded as 0 for the homozygote of the allele with the highest frequency and 2 assigned to the other homozygote. Heterozygotes were assigned values of 1. The false discovery rate was constrained to 0.05 using methodology of Benjamini and Hochberg (1995).
The designed relatedness in this project was evaluated by conducting association analyses with two random structures. The genetic covariance among animals in this project was modeled by constructing 1) the relationship matrix based on pedigree, and 2) the genomic relationship matrix based upon genotypes from the Bovine SNP50 using procedures of JMP Genomics. The pedigree-based relationship matrix was constructed with coancestry coefficients for pairs of animals according to methodology described by Falconer and Mackay (1996). There were 2,419 individuals in the pedigree. Elements of the genomic relationship were calculated as an approximation of identity-by-descent by adjusting the probability of identity-by-descent between two individuals with the average probability of identity-by-state between random individuals (Yu et al., 2006):
where n indexes genotypes, i and j index individuals with genotypes; IBDijk is identity-by-descent estimate of the probability that individuals i and j share an allele at marker genotype k that came from the same ancestor at a locus. This probability is estimated without considering pedigree information; Xik = 0, 1, 2, corresponding to genotypes at marker k in animal i; Within a genotype, p (q) is the frequency within each genotype of the nucleotide (A, C, G, or T) lower (higher) in the alphabet.
A third analysis was conducted using a completely fixed effects model with no relatedness modeled. For each of those 3 scenarios, additional analyses differed by the inclusion of family as a fixed effect (18 levels: 14 full-sibling and four half-sibling families). These six analyses were conducted on birth weight (n = 737; mean 35.0 kg; SD 5.73 kg). Subsequently, those results were used to select the model for analysis of gestation length (n = 473; mean 281 days; SD 5.39 days) and weaning weight (n = 699; mean 229.7 kg, SD 30.5 kg). All outliers (those outside the mean ± 4 SD) were excluded from analyses. Quantile-quantile plots were constructed of ranked test statistics (unadjusted probability values of F ratios for regression of phenotype on coded genotypes) at each SNP (The Wellcome Trust Case Control Consortium, 2007), and the degree of inflation of the median probability value (λ) was calculated (Devlin and Roeder, 1999) as assessment of the appropriate removal of the effects of population substructure (Voorman et al., 2011). Values of this parameter λ that are closer to 1 are favorable, and those much larger than 1 may indicate the presence of stratification that could result in spurious association of phenotype with genotype.
Locations of the genomic feature closest to each SNP locus associated in final models for each trait were determined using R statistical software and the package Map2NCBI (Hanna and Riley, 2014). Map coordinates reported are from the bovine UMD_3.1 build (Zimin et al., 2009).
Results and Discussion
Preliminary analyses indicated that a combination of year and season of birth was an efficient parameterization of these effects (p < 0.001). Cow age was retained in final models for all traits (p < 0.01), and calf gender explained significant birth weight variation. Family was considered important for all traits in these preliminary analyses (p < 0.001).
The first notable effect of refinement of the relatedness modeled was reduction of the number of detected associations (Table 1). Relative to a model that included no relatedness, the number of detected associations was reduced by over one third in analysis that modeled random animal effects with the inclusion of a pedigree-based relationship matrix. The numbers of detections when employing a genomic relationship matrix were drastically reduced. This implies that the failure to model animal as random resulted in that variation inappropriately contained in the marker regression. Smaller values of  indicated that additional refinement by employing marker relatedness as opposed to the average relatedness (with exception of parent-offspring, as that relatedness is exact) based on pedigree best accounted for that variation in the analyses (Figures S1, S2, and S3). The variation in pedigree relationships within a population may be much larger than variation within the genomic relationships among pairs with the same pedigree relationship (Hill, 2013).
indicated that additional refinement by employing marker relatedness as opposed to the average relatedness (with exception of parent-offspring, as that relatedness is exact) based on pedigree best accounted for that variation in the analyses (Figures S1, S2, and S3). The variation in pedigree relationships within a population may be much larger than variation within the genomic relationships among pairs with the same pedigree relationship (Hill, 2013).
The inclusion of the designed family fixed effect had pronounced influence on the association results. In the fixed effects model (no inclusion of relationship matrix in equations corresponding to random effects), the effect of family appeared to substitute for random structure and capture variation to the extent that this model resulted in approximately the same numbers of detections (Table 1, Figure 1) as the mixed model with pedigree-based relatedness included. The inclusion of family as a fixed effect resulted in additional associations detected in pedigree-based analyses and genomic-based analyses (Figures 2 and 3) and a slightly larger value for  in the pedigree-based analysis. This is likely because family structure is confounded with information provided in the relationship matrix. Also, there must exist non-0 covariances between levels of this effect and the relatedness modeled in these mixed models. These were modeled as 0 in these analyses, and that may also have contributed to the more liberal detections of SNP-birth weight associations.
in the pedigree-based analysis. This is likely because family structure is confounded with information provided in the relationship matrix. Also, there must exist non-0 covariances between levels of this effect and the relatedness modeled in these mixed models. These were modeled as 0 in these analyses, and that may also have contributed to the more liberal detections of SNP-birth weight associations.
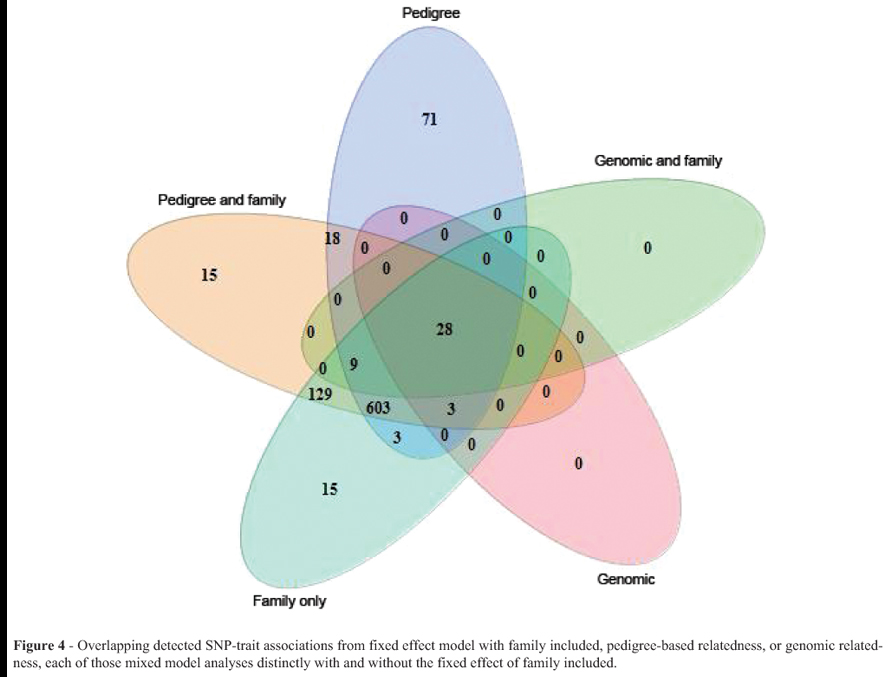
The majority of SNP loci detected as associated with birth weight in the analyses with a genomic relationship matrix included (28 of 31) were detected in each of the other analyses with different relationship parameterizations (Figure 4) except for the fixed effect model that did not include family effects. The analysis that modeled no relatedness (nor family as a fixed effect) identified a large number of loci (n = 650) that were not detected otherwise.
Zhang et al. (2006) reported high correspondence between relationship matrices constructed using pedigree and molecular information, but reported that the distribution of p values (H0: no association) from a mixed model with relationship matrix based on markers was closer to the expected distribution than those p values produced from mixed models with pedigree-based relationship matrix included or from a fixed effect model. Incorporating marker-based relationships may increase the power of association tests (Yu et al., 2006). The most recent efforts reported that the use of both pedigree and genomic relationship may be the most appropriate for prediction of breeding values (Aguilar et al., 2010; Goddard et al., 2011; Haile-Mariam et al., 2013). The appropriate modeling of relatedness was incorporation of the genomic relationship matrix without including family as a fixed effect.
Birth weight
There were 14, 8, and 9 SNP loci detected as associated with birth weight at the p < 0.001, < 0.01, and < 0.05 levels respectively (Table S1); corresponding numbers in Bos taurus studies were 35, 91, and 339 (Snelling et al., 2010), and 105, 527, and 2,095 (Lu et al., 2013). The majority of detected loci (30 of 31) were from BTA14:16.3 to 28.4 Mb. The proportion of phenotypic variance accounted for by each SNP (singly, that is, not in a multiple regression model) ranged from 0.023 to 0.067. A single locus was detected at BTA5:49.5. Most recently, Lu et al. (2013) reported multiple SNP-birth weight associations on both of these chromosomes. Kneeland et al. (2004) reported QTL for birth weight on BTA6, 19, and 21. Seven associated markers were within the bp limits of the XK, Kell blood group subunit-related family, member 4 (XKR4), and another was upstream > 25 kb of this gene (Table S1). A significant association of a SNP within this gene with birth weight was also detected in Nelore cattle (Utsunomiya et al., 2013); this was one of five total associations detected in that study, as those researchers applied Bonferroni corrections to p-values for the 434,020 loci evaluated. Detected marker-subcutaneous rump fat associations were reported for markers in and around XKR4 in Brahman and Brahman-influenced cattle (Bolormaa et al., 2011; Porto Neto et al., 2012), and this was considered a candidate gene based on marker-trait associations for average daily gain, average daily feed intake, and residual feed intake in Bos taurus cattle (Lindholm-Perry et al., 2012). Chromosome 14 has been consistently identified in association studies as important for growth traits of cattle (e.g., Snelling et al., 2010; Bolormaa et al., 2011; Lu et al., 2013); summaries of detections and genes in that region across time were detailed by Wibowo et al. (2008) and Hawken et al. (2012). Although pleiomorphic adenoma gene 1 (PLAG1) on BTA14 was considered a candidate gene for influencing hip height in cattle (Karim et al., 2011), it was not the closest gene to any SNP with detected association in the present study. The closest gene to the marker with strongest association was proenkephalin (PENK) on BTA14. This gene was identified as one of 13 found within 500 kb of the SNP most significantly associated with birth weight in Nelore cattle (Utsunomiya et al., 2013). Increasing expression of PENK in the bovine endometrium concurrent with the progression of pregnancy may indicate that this gene promotes or contributes to conceptus elongation (Forde et al., 2013).
Peters et al. (2012) reported six SNP associated with birth weight in Brangus cattle in the similar region of BTA5. Although it was not the closest feature to the detected locus on BTA5 in the present study, signal transducer and activator of transcription 6 (STAT6) is in the same region; it has been characterized with respect to SNP associations with various cattle traits (Rincón et al., 2009). Hawken et al. (2012) detected associations of SNP with body weight in a broad region of BTA5 and provided a good summary of earlier work with detections on that chromosome.
Gestation length
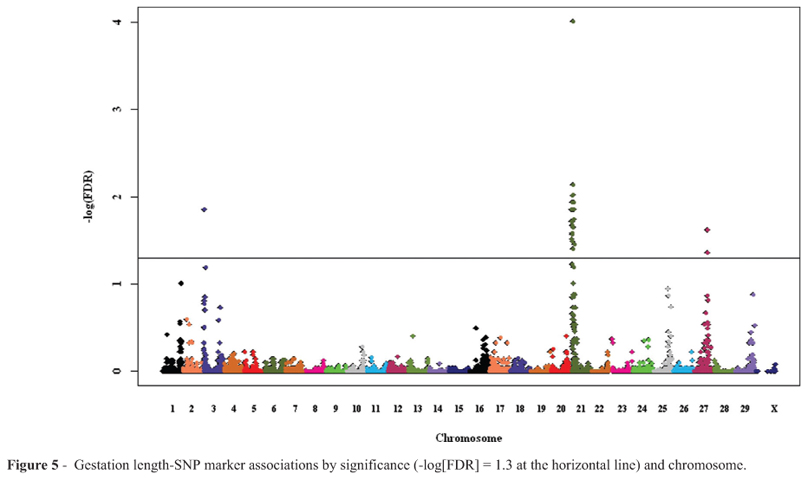
Almost all (37 of 40) of the markers associated with gestation length were on BTA21 (Figure 5; Table S2), as well as one marker on BTA3 and two on BTA27. The estimate of the degree of inflation of the median probability value ( ) was 0.98 and the quantile-quantile plot of χ2 statistics and p-values is in the supplementary information (Figure S4). These did not overlap with most markers previously detected as associated with gestation length (Maltecca et al., 2009; 2011) in dairy cattle, although Maltecca et al. (2011) identified 6 SNP on BTA3. Some SNP in imprinted domain regions on BTA13 were associated with calving difficulty and gestation length of dairy cows (Sikora et al., 2011). A similar region was identified on BTA21 (beginning at BTA21:68.1 Mb; Magee et al., 2010) including delta-like 1 homolog DLK1 and deiodinase, iodothyronine, type III (DIO3); this region was associated with various traits in dairy cattle, including gestation length (Magee et al., 2011). The closest two associated markers in the present study were approximately 30 Mb upstream of this region. Eighteen associated markers were within the bp boundaries of eight genes and four pseudogenes (Table S2).
) was 0.98 and the quantile-quantile plot of χ2 statistics and p-values is in the supplementary information (Figure S4). These did not overlap with most markers previously detected as associated with gestation length (Maltecca et al., 2009; 2011) in dairy cattle, although Maltecca et al. (2011) identified 6 SNP on BTA3. Some SNP in imprinted domain regions on BTA13 were associated with calving difficulty and gestation length of dairy cows (Sikora et al., 2011). A similar region was identified on BTA21 (beginning at BTA21:68.1 Mb; Magee et al., 2010) including delta-like 1 homolog DLK1 and deiodinase, iodothyronine, type III (DIO3); this region was associated with various traits in dairy cattle, including gestation length (Magee et al., 2011). The closest two associated markers in the present study were approximately 30 Mb upstream of this region. Eighteen associated markers were within the bp boundaries of eight genes and four pseudogenes (Table S2).
Weaning weight
All but one of the 26 detected SNP-weaning weight associations (Figure 6; Table S3) were on BTA14. The quantile-quantile plot of χ2 statistics and p-values is in the supplementary information (Figure S4);  was 0.99. Most of those (n = 21) were also detected as associated with birth weight; this result supported the conclusions of Snelling et al. (2010), that is, genomic regions responsible for variation in both traits were largely the same and that using genomic information to select for growth in one phase while limiting or minimizing growth in another phase would be difficult. A single locus at BTA10:26.6 Mb was detected as associated with weaning weight. Lu et al. (2013) detected associations with weaning weight on these chromosomes (plus others) in similar regions. Snelling et al. (2010) detected associations of the direct component of weaning weight with SNP on BTA10 and BTA14, but primarily on BTA6; they detected maternal components of weaning weight associated with SNP on the chromosomes detected in the present study. It should be noted that these mentioned represent a small subset of the overall detections from Snelling et al. (2010) that spanned essentially all chromosomes. Peters et al. (2012) reported a window of SNP associated with 205-d weight in Brangus heifers on BTA10. Fifteen of the SNP with detected association with weaning weight were located within physical boundaries of eight genes and one pseudogene (Table S3).
was 0.99. Most of those (n = 21) were also detected as associated with birth weight; this result supported the conclusions of Snelling et al. (2010), that is, genomic regions responsible for variation in both traits were largely the same and that using genomic information to select for growth in one phase while limiting or minimizing growth in another phase would be difficult. A single locus at BTA10:26.6 Mb was detected as associated with weaning weight. Lu et al. (2013) detected associations with weaning weight on these chromosomes (plus others) in similar regions. Snelling et al. (2010) detected associations of the direct component of weaning weight with SNP on BTA10 and BTA14, but primarily on BTA6; they detected maternal components of weaning weight associated with SNP on the chromosomes detected in the present study. It should be noted that these mentioned represent a small subset of the overall detections from Snelling et al. (2010) that spanned essentially all chromosomes. Peters et al. (2012) reported a window of SNP associated with 205-d weight in Brangus heifers on BTA10. Fifteen of the SNP with detected association with weaning weight were located within physical boundaries of eight genes and one pseudogene (Table S3).
Conclusions and Implications
In mixed model association analyses, marker-based relationship appeared to best capture trait variance corresponding to relatedness. In the absence of either a relationship matrix based on marker genotypes or pedigree, the fixed effect of family appeared to assume that role, but to a lesser extent. Detected associations increased when more primitive parameterizations of relatedness (or no relatedness) were employed, apparently because of stratification. Inclusion of family as a fixed effect in addition to either relationship matrix also resulted in increased associations detected, either due to duplicative modeling of relatedness or due to inappropriate assumption of non-zero covariance between fixed effect levels and random effect levels. Detected associations with birth weight and weaning weight were primarily on BTA14 and consistent with other reports. The strongest association for birth weight was for a marker within PENK, which has been identified as expressed in placenta. Detected associations with gestation length were primarily on BTA21.
Acknowledgments
This project was supported by National Research Initiative Competitive Grant no. 2008-35205-18767 from the USDA National Institute of Food and Agriculture. Appreciation is noted for B. D. Johnson, D. A. Gann, and all personnel at the Texas A&M AgriLife Research Center at McGregor.
Supplementary Material
The following online material is available for this article:
Table S1 - Single nucleotide polymorphism markers associated with birth weight.
Table S2 - Single nucleotide polymorphism markers associated with gestation length.
Table S3 - Single nucleotide polymorphism markers associated with weaning weight.
Figure S1 - Quantile-quantile plots and 95% confidence bands of observed χ2 values against expected probability values of birth weight-marker association F statistics from models with no relationship matrix included, and the fixed effect family included (upper) or not included (lower).
Figure S2- Quantile-quantile plots for models based on average pedigree relatedness.
Figure S3- Quantile-quantile plots for models with genomic relationship matrix.
Figure S4- Quantile-quantile plots of gestation length-marker association and weaning weight- marker association F statistics.
This material is available as part of the online article from http://www.scielo.br/gmb.
Data Access: Data will be shared with collaborators upon request.
Received: March 7, 2014;
Accepted: June 12, 2014.
Associate Editor: Dario Grattapaglia
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S and Lawlor TJ (2010) A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci 93:743-752.
- Benjamini Y and Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc B 57:289-300.
- Bolormaa S, Porto Neto LR, Zhang YD, Bunch RJ, Harrison BE, Goddard ME and Barendse W (2011) A genome-wide association study of meat and carcass traits in Australian cattle. J Anim Sci 89:2297-2309.
- Devlin B and Roeder K (1999) Genomic control for association studies. Biometrics 55:997-1004.
- Falconer DS and Mackay, TFC (1996) Introduction to Quantitative Genetics. 4th edition. Pearson Educ. Ltd., Essex, 480 pp.
- Forde N, Mehta JP, McGettigan PA, Mamo S, Bazer FW, Spencer TE and Lonergan P (2013) Alterations in expression of endometrial genes coding for proteins secreted into the uterine lumen during conceptus elongation in cattle. BMC Genomics 14:e321.
- Goddard ME, Hayes BJ and Meuwissen THE (2011) Using the genomic relationship matrix to predict the accuracy of genomic selection. J Anim Breed Genet 128:409-421.
- Haile-Mariam M, Nieuwhof GJ, Beard KT, Konstatinov KV and Hayes BJ (2013) Comparison of heritabilities of dairy traits in Australian Holstein-Friesian cattle from genomic and pedigree data and implications for genomic evaluations. J Anim Breed Genet 130:20-31.
- Hanna LLH and Riley DG (2014) Mapping genomic markers to closest feature using the R package Map2NCBI. Livest Sci 162:59-65.
- Hawken RJ, Zhang YD, Fortes MRS, Collis E, Barris WC, Corbet NJ, Williams PJ, Fordyce G, Holroyd RG, Walkley JRW, et al. (2012) Genome-wide association studies of female reproduction in tropically adapted cattle. J Anim Sci 90:1398-1410.
- Hayes BJ and Goddard ME (2008) Prediction of breeding values using marker-derived relationship matrices. J Anim Sci 86:2089-2091.
- Henderson CR (1963) Selection index and expected genetic advance. In: Statistical Genetics and Plant Breeding. Nat Acad Sci National Research Council, Publ. No. 982, Washington, DC, pp 141-163.
- Hill WG (2013) On estimation of genetic variance within families using genome-wide identity-by-descent sharing. Genet Sel Evol 45:32.
- Hong H, Xu L, Liu J, Jones WD, Su Z, Ning B, Perkins R, Ge W, Miclaus K, Zhang L, et al. (2012) Technical reproducibility of genotyping SNP arrays used in genome-wide association studies. PLoS One 7:e44483.
- Karim L, Takeda H, Lin L, Druet T, Arias JAC, Baurain D, Cambisano N, Davis SR, Farnir F, Grisart B, et al. (2011) Variants modulating the expression of a chomrosome domain encompassing PLAG1 influence bovine stature. Nat Genet 43:405-413.
- Kneeland J, Li C, Basarab J, Snelling WM, Benkel B, Murdoch B, Hansen C and Moore SS (2004) Identification and fine mapping of quantitative trait loci for growth traits on bovine chromosomes 2, 6, 14, 19, 21, and 23 within one commercial line of Bos taurus J Anim Sci 82:3405-3414.
- Lindholm-Perry AK, Kuehn LA, Smith TPL, Ferrell CL, Jenkins TG, Freetly HC and Snelling WM (2012) A region on BTA14 that includes the positional candidate genes LYPLA1, XKR4 and TMEM68 is associated with feed intake and growth phenotypes in cattle. Anim Genet 43:216-219.
- Liu Y, Jansen GB and Lin CY (2002) The covariance between relatives conditional on genetic markers. Gen Sel Evol 34:657-678.
- Lu D, Miller S, Sargolzaei M, Kelly M, Vander Voot G, Caldwell T, Plastow G and Moore S (2013) Genome-wide association analyses for growth and feed efficiency traits in beef cattle. J Anim Sci 91:3612-3633.
- Magee DA, Berry DP, Berkowicz EW, Sikora KM, Howard DJ, Mullen MP, Evans RD, Spillane C and MacHugh DE (2011) Single nucleotide polymorphisms within the bovine DKLI-DIO3 imprinted domain are associated with economically important production traits in cattle. J Hered 102:94-101.
- Magee DA, Sikora KM, Berkowicz EW, Berry DP, Howard DJ, Mullen MP, Evans RD, Spillane C and MacHugh DE (2010) DNA sequence polymorphisms in a panel of eight candidate bovine imprinted genes and their association with performance traits in Irish Holstein-Friesian cattle. BMC Genet 11:e93.
- Maltecca C, Weigel KA, Khatib H, Cowan M and Bagnato A (2009) Whole-genome scan for quantitative trait loci associated with birth weight, gestation length and passive immune transfer in a Holstein x Jersey crossbred population. Anim Genet 40:27-34.
- Maltecca C, Gray KA, Weigel KA, Cassady JP and Ashwell M (2011) A genome-wide association study of direct gestation length in US Holstein and Italian Brown populations. Anim Genet 42:585-591.
- Peters SO, Kizilkaya K, Garrick DJ, Fernando RL, Reecy JM, Weaber RL, Silver GA and Thomas MG (2012) Bayesian genome-wide association analysis of growth and yearling ultrasound measures of carcass traits in Brangus heifers. J Anim Sci 90:3398-3409.
- Porto Neto LR, Bunch RJ, Harrison BE and Barendse W (2012) Variation in the XKR4 gene was significantly associated with subcutaneous rump fat thickness in indicine and composite cattle. Anim Genet 43:785-789.
- Riley DG, Welsh Jr. TH, Gill CA, Hulsman LL, Herring AD, Riggs PK, Sawyer JE and Sanders JO (2013) Whole genome association of SNP with newborn calf cannon bone length. Livest Sci 155:186-196.
- Rincón G, Farber EA, Farber CR, Nkrumah JD and Medrano JF (2009) Polymorphisms in the STAT6 gene and their association with carcass traits in feedlot cattle. Anim Genet 40:878-882.
- Sham PC, Cherny SS and Purcell S (2009) Application of genome-wide SNP data for uncovering pairwise relationships and quantitative loci. Genetica 136:237-243.
- Sikora KM, Magee DA, Berkowicz EW, Berry DP, Howard DJ, Mullen MP, Evans RD, MacHugh DE and Spillane C (2011) DNA sequence polymorphisms within the bovine guanine nucleotide-binding protein Gs subunit alpha (Gsα)-encoding (GNAS) genomic imprinting domain are associated with performance traits. BMC Genet 12:e4.
- Snelling WM, Allan MF, Keele JW, Kuehn LA, McDaneld T, Smith TPL, Sonstegard TS, Thallman RM and Bennett GL (2010) Genome-wide association study of growth in crossbred beef cattle. J Anim Sci 88:837-848.
- The Wellcome Trust Case Control Consortium (2007) Genome-wide association of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661-678.
- Thomas SC (2005) The estimation of genetic relationships using molecular markers and their efficiency in estimating heritability in natural populations. (Special issue. Population genetics, quantitative genetics and animal improvement. Papers in honour of William [Bill] Hill). Philos Trans R Soc B Biol Sci 360:1457-1467.
- Utsunomiya YT, do Carmo AS, Carvalheiro R, Neves HHR, Matos MC, Zavarez LB, Pérez O'Brien AM, Sölkner J, McEwan JC, et al. (2013) Genome-wide association study for birth weight in Nellore cattle points to previously described orthologous genes affecting human and bovine height. BMC Genet 14:e52.
- Visscher PM (2009) Whole genome approaches to quantitative genetics. Genetica 136:351-358.
- Visscher PM, Medland SE, Ferreira MA, Morley KI, Zhu G, Cornes BK, Montgomery GW and Martin NG (2006) Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet 2:e41.
- Voorman A, Lumley T, McKnight B and Rice K (2011) Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS One 6:e19416.
- Wigginton JE, Cutler DJ and Abecasis, GR (2005) A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 76:887-893.
- Wibowo TA, Gaskins CT, Newberry RC, Thorgaard GH, Michal JJ and Jiang Z (2008) Genome assembly anchored QTL map of bovine chromosome 14. Int J Biol Sci 4:406-414.
- Wolc A, Arango J, Settar P, Fulton JE, O'Sullivan NP, Preisinger R, Fernando R, Garrick DJ and Dekkers JCM (2013) Analysis of egg production in layer chickens using a random regression model with genomic relationships. Poultry Sci 92:1486-1491.
- Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gault BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203-208.
- Zhang Z, Mateescu RG, Lust G, Phavaphutanon J, Tsai K, Murphy K, Todhunter RJ and Buckler ES (2006) Association mapping accounting for background QTLs: Relationship based on pedigree vs. molecular markers. Proc 8th World Congr Genet Appl Livest Prod 20-08, Belo Horizonte, CD-ROM Communication No. 20-08.
- Zhang Z, Todhunter RJ, Buckler ES and Van Vleck LD (2007) Use of marker-based relationships with multiple-trait derivative-free restricted maximal likelihood. J Anim Sci 85:881-885.
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, et al. (2009) A whole-genome assembly of the domestic cow, Bos taurus Genome Biol 10:R42.
Publication Dates
-
Publication in this collection
22 Sept 2014 -
Date of issue
Sept 2014
History
-
Received
07 Mar 2014 -
Accepted
12 June 2014

 Alternative parameterizations of relatedness in whole genome association analysis of pre-weaning traits of Nelore-Angus calves
Alternative parameterizations of relatedness in whole genome association analysis of pre-weaning traits of Nelore-Angus calves