Abstract
The earliest Cape Muslims were brought to the Cape (Cape Town -South Africa) from Africa and Asia from 1652 to 1834. They were part of an involuntary migration of slaves, political prisoners and convicts, and they contributed to the ethnic diversity of the present Cape Muslim population of South Africa. The history of the Cape Muslims has been well documented and researched however no in-depth genetic studies have been undertaken. The aim of the present study was to determine the respective African, Asian and European contributions to the mtDNA (maternal) and Y-chromosomal (paternal) gene pool of the Cape Muslim population, by analyzing DNA samples of 100 unrelated Muslim males born in the Cape Metropolitan area. A panel of six mtDNA and eight Y-chromosome SNP markers were screened using polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP). Overall admixture estimates for the maternal line indicated Asian (0.4168) and African mtDNA (0.4005) as the main contributors. The admixture estimates for the paternal line, however, showed a predominance of the Asian contribution (0.7852). The findings are in accordance with historical data on the origins of the early Cape Muslims.
PCR-RFLP; genetic polymorphism; mitochondrial DNA; population genetic structure; chromosome variations
Reconstruction of major maternal and paternal lineages of the Cape Muslim population
Shafieka Isaacs; Tasneem Geduld-Ullah; Mongi Benjeddou
Department of Biotechnology, University of the Western Cape, Bellville, Cape Town, South Africa
ABSTRACT
The earliest Cape Muslims were brought to the Cape (Cape Town -South Africa) from Africa and Asia from 1652 to 1834. They were part of an involuntary migration of slaves, political prisoners and convicts, and they contributed to the ethnic diversity of the present Cape Muslim population of South Africa. The history of the Cape Muslims has been well documented and researched however no in-depth genetic studies have been undertaken. The aim of the present study was to determine the respective African, Asian and European contributions to the mtDNA (maternal) and Y-chromosomal (paternal) gene pool of the Cape Muslim population, by analyzing DNA samples of 100 unrelated Muslim males born in the Cape Metropolitan area. A panel of six mtDNA and eight Y-chromosome SNP markers were screened using polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP). Overall admixture estimates for the maternal line indicated Asian (0.4168) and African mtDNA (0.4005) as the main contributors. The admixture estimates for the paternal line, however, showed a predominance of the Asian contribution (0.7852). The findings are in accordance with historical data on the origins of the early Cape Muslims.
Keywords: PCR-RFLP, genetic polymorphism, mitochondrial DNA, population genetic structure, chromosome variations.
Introduction
In 1652, the first Dutch settlers arrived in Cape Town, South Africa, to establish a refreshment station for the Dutch East India Company (DEIC) ships en route to and from the East (Du Pre, 1994). The importation of slaves shortly began after the establishment of the refreshment station serving as the major labour force at the Cape, following failed attempts to enslave the indigenous Khoisan people (Da Costa and Davids, 1994; Shell, 2000). The DEIC imported slaves from the East Coast of Africa, India, Madagascar, Sri Lanka and the Indonesian Archipelago. (Bradlow and Cairns, 1978; Davids, 1980; Mahida, 1993; Da Costa and Davids, 1994; Shell, 2000). The importation of slaves remained a common practice at the Cape even after the transfer of the colony to British Empire in 1795 and again in 1806 (Giliomee, 2003). Alongside the slaves, convicts and political exiles (mainly from Indonesia) were also sent to the Cape, some of whom were prominent Muslim clerics (adherents of the faith Islam) who later became known as the Vryezwarten or Free Blacks (Bradlow and Cairns, 1978; Davids, 1980; Mahida, 1993; Shell, 2000). It is estimated that 63,000 slaves were brought to South Africa between 1652 and 1807, when the British abolished the oceanic slave trade (Shell, 2000). The major proportion of slaves, however, was not imported but were Cape-born (Bradlow and Cairns, 1978). These slaves were of mixed parentage due to the mixed relations among early White settlers, the Khoisan, slaves, and the so-called Bantuspeaking people (Pickel, 1997). In the early 18th century the slave population exceeded the settler population, with males outweighing the number of females in both the slave and free populations (Shell, 2000). Marriage between European men and women, who were either Khoisan, manumitted (freed) slaves or of mixed parentage (Keegan, 1996), and between Khoisan and slaves were thus not uncommon and were socially acceptable, but already from the 18th century, offspring of such mixed marriages and liaisons mainly were assimilated into the growing population group known as 'Coloured', after race-based restrictions were imposed by the British (Mountain, 2003). These periods also proved extremely fertile for the spread of Islam, which was mainly attributed to the growing number of Free Blacks who were well schooled in Islam and eager to convert slaves. Furthermore, Dutch colonist encouraged slaves to embrace Islam fearing the loss of their property after a Placaaten (Law) was issued that prevented the sale of baptised Christian slaves. Islam became a flourishing religion after the emancipation of slaves in 1834, and by 1840 they represented a third of the total Colony population, which had then become a more cohesive group later to be known as Cape Malay (Davids, 1980). This distinct ethnic identity was later re-enforced by the apartheid government Population Registration Act of 1950, which classified Cape Malay as a subgroup of the Coloured classification (Da Costa Y 1990, PhD thesis. University of South Africa, Pretoria). The term Malay was initially introduced by whites to describe Muslims and subsequently became synonymous with being Muslim (Bickford-Smith, 1994). Terms such as Coloured Muslim, Javaan, and Mohammedan were also used to reflect the religious and diverse ethnic identity of Muslims, later to be used interchangeably by Muslims themselves (Adihikari, 1989). In recent times the term Cape Muslim has become a more socially acceptable term to describe the Muslim community residing in the Cape Metropolitan region, which consists of diverse ethnic subgroups the majority being the descendants of early Cape Muslims (who mainly self-identify as Cape Coloured Muslim and Cape Malay Muslim) and the descendants of Indian immigrants (Cape Indian Muslims). The history of the Cape Muslims has been well documented and researched, however no significant genetic studies have been undertaken. To date only one genetic study using Y-STR markers has attempted to analyze the parental contributions made to this community (Cloete et al., 2010). In the study, nine Y-STR loci (DYS19, DYS389-I, DYS389-II, DYS390, DYS391, DYS392, DYS393) were used to characterize the minimal haplotype, and the duplicated locus DYS385) as well as eight widely used loci of DYS449, DYS481, DYS518, DYS557, DYS570, DYS607, DYS612 and DYS614. Factorial Correspondence Analysis was used to establish genetic relations between the Cape Muslim population using published data from three South African populations namely Asian Indian, European English and Xhosa. The results indicated that the Cape Muslim population was more closely related to the Asian Indian population, followed by the European English in the second place, and more distantly related to the Xhosa (Cloete et al., 2010). In the present study, mitochondrial DNA (mtDNA) and the nonrecombining Y chromosome (NRY) were screened using PCR-RFLP analysis with the aim of determining the African, Asian and European contributions to the mtDNA (maternal) and NRY (paternal) of the Cape Muslim population, thereby providing some insight into the genetic ancestry of this population.
Material and Methods
Sample collection and DNA extraction
Buccal swab samples were collected from a 100 anonymous, unrelated Muslim males born in the Cape Town Metropolitan area. Information pertaining to donor's languages spoken, familial birthplaces, and a short genealogy of two generations for maternal and paternal family members to establish regional ancestry were also obtained. This was done to draw more conclusive answers regarding the population history and explore past demographic events of this population. The self-perceived ethnic identity listed by donors and the number of individuals in each Cape Muslim subgroup were: Cape Coloured Muslim (n = 34), Cape Indian Muslim (n = 29), Cape Malay Muslim (n = 27) and Cape Other Muslim (n = 10). The term 'Other' refers to individuals that were reluctant to indicate an ethnicity or formed part of a Cape Muslim minority subgroup. The question of identity in the Cape Muslim community has always been a point of interest. A study conducted by Da Costa (1994) considered the problem of identity amongst Muslims, post-apartheid. His research suggested that many Muslims chose a religious identity over other forms of identity such as national origin or ethnicity. All samples were obtained with informed consent, and ethical clearance for the study was approved by the Senate Research Committee of the University of the Western Cape, South Africa. DNA was extracted from buccal swabs using the BuccalAmpTM DNA extraction kit (Epicentre Technologies) according to the manufacturer's instructions. Extracted DNA was further purified with a standard phenol chloroform protocol to improve the quality of the DNA as well as the sensitivity of the PCR.
Mitochondrial DNA
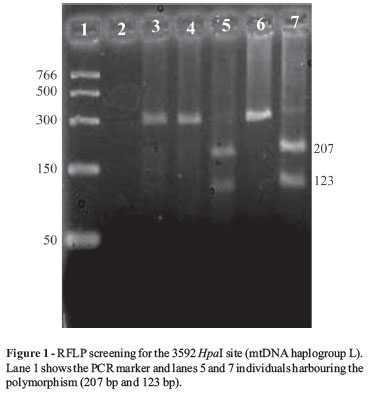
A total of six restriction sites and one insertion/deletion polymorphism (IDP) were analyzed in the study according to published PCR-RFLP protocols. The markers included an AluI loss at np 7025 (denotes haplogroup H), BstOI loss at np 13704 (denotes haplogroup J), HpaI gain at np 3592 (denotes haplogroup L), DdeI gain at np10394 and AluI gain at np 10397 (denotes haplogroup M) (Santos et al., 2004). The IDP screened was the COII/tRNALys intergenic 9-bp deletion (denotes haplogroup B) (Martinez-Cruzado et al., 2001) which has routinely been used to assess Asian and Asian-derived origins in populations (Edwin et al., 2002). This was done following a hierarchical approach to differentiate haplogroups firstly by their DdeI 10394/AluI 10397 motif (designated as +/+, +/-and -/-) then testing for the haplogroup specific marker reducing the potential to which an unknown mtDNA might belong.
Y-Chromosome
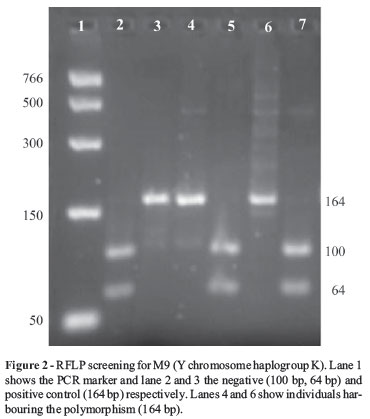
Y-chromosomal binary markers consisting of seven SNPs: M2, M35, M52, M170, M9, M175, M173 and one Alu insertion/deletion (YAP) polymorphism were used to screen DNA samples. The markers allowed the identification, of haplogroups E1b1a, E1b1b1, H1, I, K, O, R1 and DE according to methods described by (Hammer and Horai, 1995; Underhill et al., 1997, 2000; Flores et al., 2003; Kayser et al., 2005) with only minor modifications. The nomenclature used to assign haplogroups was done according to the Y Chromosome Consortium, 2002.
Restriction digestion
The restriction digestion reactions were performed using 10 µL of the unpurified amplified product, incubated with the haplogroup specific restriction enzyme according to conditions specified by the manufacturer. The digested fragments were run alongside an undigested simile in a 3% agarose gel, previously stained with ethidium bromide and photodocumentated under UV light.
Statistical analyses
The frequencies and the number of mtDNA and NRY haplogroups were calculated by direct counting, and the admixture proportions of the maternal (mtDNA) and paternal (NRY) lines were estimated by ADMIX 95 software based on the gene identity method (Chakraborty, 1985). Principal component analysis (PCA) was done based on haplogroup frequencies using the XLSTAT program implemented in Excel (Agresti, 1990). The populations hypothesized to have contributed to the Cape Muslim admixture were chosen from published data representing three major groups namely (1) African (Khoisan), (2) Asian (Indian and South-East populations) and (3) European. Sample sizes of African, Asian and European populations were respectivelyn=74 (Chen et al., 2000), n = 378 (Kivisild et al., 1999; Sudoyo et al., 2002), and n = 956 (Byrne et al., 2008) for mtDNA, and n = 129 (Underhill et al., 2001, Cruciani et al., 2002), n = 398 (Kayser et al., 2003; Cordaux et al., 2004; Tajima et al., 2004), andn=66 (Semino et al., 2000) for NRY.
Results
PCR-RFLP analysis
Most of the markers used in the study were screened using PCR-RFLP analysis (Figure 1) with the exception of the 9 bp deletion for haplogroup B and the YAP element defining haplogroup DE. These markers were characterized by either an insertion or deletion polymorphism thereby rendering restriction digestion non-essential. The banding pattern of the amplified fragment was therefore sufficient to discern a genotype. None of the donors harboured the 9 bp deletion while three individuals harboured the YAP element. Haplogroup markers defined by the absence of cleaved digested fragments were verified using both positive and negative controls (Figure 2).
Maternal lineages of the Cape Muslims
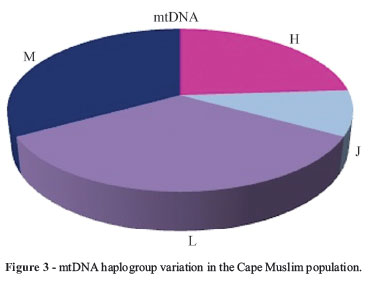
The mtDNA haplogroups observed in the study are reported in Table 1. Their ancestry has phylogenetically been established (Torroni et al., 1996, 2006; Wallace et al., 1999; Van Oven and Kayser, 2008). Cape Muslims maternal lineages mainly belonged to haplogroups L and M (Figure 3). Haplogroup L is an African specific lineage that is distributed throughout the African continent with an increased frequency in Sub-Saharan African populations (Mishmar et al., 2003; Underhill and Kivisild, 2007; Maji et al., 2009).
The haplogroup distribution differed among the selfperceived subgroups, with Cape Coloured Muslims (47%) and Cape Malay Muslims (44%) showing the highest frequencies while Cape Other Muslims (20%) and Cape Indian Muslims (14%) had the lowest. The Cape Indian Muslims' mtDNA mainly belonged to an Asian lineage, haplogroup M which has the most frequent and diverse distribution in Asia, Melanesia and Native American populations (Kivisild et al., 1999; Bermisheva et al., 2002; Maji et al., 2009). Furthermore, it represents the most dominant mtDNA haplogroup in India (Kivisild et al., 1999; Roychoudhury et al., 2000; Schurr and Wallace, 2002; Edwin et al., 2002; Cordaux et al., 2003; Tripathy et al., 2008, Maji et al., 2009).
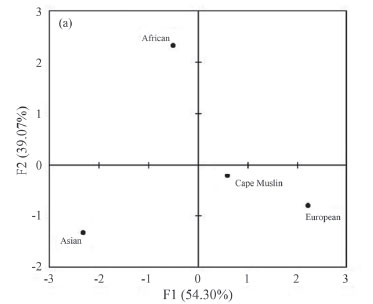
The European mtDNA lineages H and J were both observed in the study albeit at different frequencies. Haplogroup H was the third most frequent mtDNA haplogroup (24%) in the study. Cape Other Muslim displayed the highest frequency (50%) followed by Cape Indian Muslims (38%). Haplogroup H is the most prevalent mtDNA in Europe with the highest frequency (40-50%) in western and northern Europe. In southern, southwestern and eastern Europe, North Africa and Turkey an intermediate frequency (20-40%) is observed. The lowest frequency for haplo-group H is reported in the Middle East, India, and Siberia (Wallace et al., 1999; Alves-Silva et al., 2000; Bermisheva et al., 2002; Cordaux et al., 2003). Haplogroup J was mainly prevalent among the Cape Malay Muslims (15%) and Cape Coloured Muslims (12%). Cape Indian Muslims had the lowest frequency (3%), while Cape Other Muslims harboured no J mtDNA. Haplogroup J represents 11.3% of European mtDNA (Wallace et al., 1999) with an appreciable frequency in the Near East (Wells, 2007). The admixture estimates for the maternal line (Table 2) indicate Asian (0.4168) and African mtDNA (0.4005) as the main contributors. The PCA plot for Cape Muslim subgroups (Figure 5b) corresponded with these finding. However when the population was analysed as a group, the Cape Muslims (Figure 5a) clustered with Europeans. This could possibly be due to Western Eurasian haplogroups (H and J) being shared among Indian and European populations.
Paternal lineages of the Cape Muslims
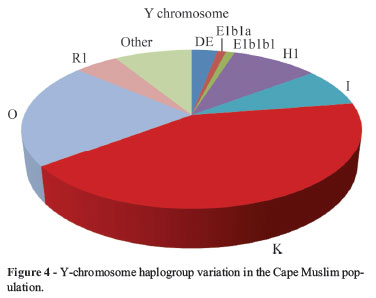
Cape Muslim paternal lineages were classified into eight different haplogroups (Figure 4) mainly belonging to haplogroups H1, K and O (Table 1), with only a small percentage of paternal lineages remaining unresolved. Cape Malay Muslims displayed the highest frequency (52%) for haplogroup K, while Cape Indian Muslims demonstrated a high frequency for haplogroup O (38%) and H1 (17%). Haplogroup K is distributed throughout Asia having a substantial presence in East Asia (Basu et al.; 2003; Kumar et al., 2007). Haplogroup O characterises a major Y-chromosome lineage in East Asia and represents 80-90% of Y-chromosomes in Southeast and East Asia, with high frequencies observed in Indian populations (Underhill, 2004; Wells, 2007; Debnath et al., 2011). Haplogroup H1 is restricted to India and is frequently found among tribal and lower caste population groups (Krithika et al., 2007; Karafet et al., 2008).
The European lineages observed in the study were haplogroups R1 and I. These haplogroups are widespread across Europe and are found at high frequencies in western Europe (Cavalli-Sforza and Feldman, 2003; Rootsi et al., 2004; Fregel et al., 2009). Haplogroup I was the most frequent in the study it observes its highest frequency in Cape Coloured Muslims (17%), while R1 in Cape Other Muslims (10%). These haplogroups were however, absent in Cape Indian Muslims.
Haplogroups DE, E1b1a and E1b1b1 make up the remaining paternal lineages. These haplogroups had the lowest frequencies in the study and are mainly prevalent among African populations (Underhill et al., 2001). The YAP element defining haplogroup DE was observed only in Cape Coloured Muslims (6%) and Cape Other Muslims (10%). This haplogroup defines both Asian (haplogroup D) and African (haplogroup E) Y-chromosome lineages. Nonetheless, its greatest frequency is observed in Sub Saharan populations (Hammer and Horai, 1995; Agrawal et al., 2005). Haplogroup E1b1a is common in Sub Saharan populations and is used to trace Bantu migrations (Hammer and Zegura 2002; Beleza et al., 2005; Fregel et al., 2009). The haplogroup was observed in a Cape Indian Muslim while haplogroup E1b1b1 was present in a Cape Other Muslim. The haplogroup is frequent among East and North African populations (Hammer and Zegura, 2002; Beleza et al., 2005). Estimates for admixture through the paternal lineage (Table 3) indicate a predominance of the Asian contribution (0.7852), which is reflected in the PCA analysis of the Cape Muslims (Figure 6a) and the self-perceived subgroups (Figure 6b).
Discussion
The data from the mtDNA haplogroups (Figure 3) and admixture estimates (Table 2) suggest a strong African contribution to the maternal gene pool of the Cape Muslim population, particularly for the Cape Coloured Muslim and Cape Malay Muslim. Cape Other Muslim (20%) and Cape Indian Muslim (14%) showed a reduced frequency (Table 1).
Haplogroup L is widely distributed among African populations especially in Sub-Saharan Africa (Wallace et al., 1999; Gonder et al., 2007). The presence of this haplogroup suggests Khoisan and Bantu females as likely contributors of this lineage. However, African slaves originating from Mozambique and Madagascar may also have been contributors, as haplogroup L represents a common mtDNA among the Malagasy population (Hurles et al., 2005). In Cape Indian Muslims, however, haplogroup L accounts for 14% of their mtDNA, which could possibly be attributed to geneflow with the Siddis, as the genealogical survey indicated that maternal grandmothers were recent immigrants from India. The Siddis are tribal groups in India that were brought as slaves during the 16th and 17th century to India from East Africa and other regions in Africa such as Mozambique (Lodhi 1992; Gauniyal et al., 2008; Shah et al., 2011). Analysis of the genealogical survey indicated that maternal grandmothers were recent immigrants from India. These results also suggest that the remaining L mtDNA was mainly derived through recent admixture with either the Cape Coloured Muslim or Cape Malay Muslim.
Haplogroup M was the second most frequent haplogroup in the study. It is common among Asian populations and is the most dominant mtDNA haplogroup in India (Kivisild et al., 1999; Roychoudhury et al., 2000; Edwin et al., 2002; Schurr and Wallace 2002; Cordaux et al., 2003; Tripathy et al., 2008, Maji et al., 2009). A study by Cordaux et al. (2003) reported the haplogroup's frequency among northeastern tribes as being 51% while eastern, central and southern tribes had an equal share in the frequency of 76%. This result was consistent with findings in our study as the Cape Indian Muslims displayed the highest frequency (45%) for haplogroup M. A moderate frequency for haplogroup M was observed among the Cape Coloured Muslims (32%), Cape Malay Muslims (22%), and Cape Other Muslims (30%). The origin of their mtDNA was likely mainly derived from female slaves originating from India and South East Asia, as a significant number of Asian slaves originated from India and the Indonesian Archipelago (Bradlow and Cairns, 1978; Davids, 1980; Da Costa and Davids, 1994). Haplogroup M may even have been introduced via slaves from Madagascar due to their mixed Indonesian ancestry and recently through admixture with Indian and other Asian populations (Hurles et al., 2005).
Haplogroup H was the third most frequent mtDNA (24%) in our study. In the self-perceived subgroups, Cape Other Muslim displayed the highest frequency (50%) followed by Cape Indian Muslim (38%). A lower frequency for haplogroup J was observed, this being mainly prevalent among the Cape Malay Muslim (15%) and Cape Coloured Muslim (12%). The Cape Indian Muslim had the lowest frequency (3%), while the Cape Other Muslim harboured no J mtDNA. Haplogroup H and J may have been introduced via recent and historical admixture with European and Indian populations, as haplogroup H represents nearly 50% of European mtDna, while haplogroup J represents 11.3% of European mtDNA (Torroni et al., 1996; Lell and Wallace, 2000; Bermisheva et al., 2002; Loogväli et al., 2004; Piechota et al., 2004; Alzualde et al., 2005; Manwaring et al., 2006). Furthermore, West Eurasian haplogroups make up 20% to 30% of Indian mtDNA, with northern Indians possessing a higher frequency of West Eurasian mtDNA than southern Indians (Roychoudury et al., 2000; Tripathy et al., 2008).
Haplogroup B was the only mtDNA not observed in the study. The 9 bp deletion defining this haplogroup is routinely used to infer Asian and Polynesian ancestry (Ballinger et al., 1992; Stone and Stoneking, 1998; Edwin et al., 2002; Berniell-Lee et al., 2008). Haplogroup B has a high to moderate frequency among East Indonesians and Malays (Stone and Stoneking, 1998; Merriwether et al., 1999; Schurr and Wallace, 2002). Therefore, the absence of this haplogroup was unexpected, as a number of slaves had originated from the Indonesian Archipelago (Bradlow and Cairns, 1978; Davids, 1980; Da Costa and Davids, 1994).
To further characterise the genetic variability of the Cape Muslim population and to analyse their maternal origin in greater detail, admixture analysis and PCA analyses were carried out. Table 3 indicates the African, Asian and European maternal ancestry present in the Cape Muslim population and subgroups. Admixture estimates indicated that the Cape Muslim mtDNA is mainly of African and less so Asian and European ancestry. The PCA analysis (Figure 4) indicated that Cape Coloured Muslim and Cape Malay Muslim are more closely related to African populations, while the Cape Indian Muslim and Cape Other Muslim are so to Indian populations.
A comparison of mtDNA and NRY lineages present in the Coloured population of the Western Cape
A number of researchers have recently investigated the mixed ancestry of the Coloured population in the Western Cape (Tishkoff et al., 2009; De Wit et al., 2010; Patterson et al., 2010; Quintana-Murci et al., 2010). This is largely due to the unique opportunities this population provides for studies related to population history, natural selection, and admixture mapping. The study by Tishkoff et al. (2009) analyzed 1,327 nuclear microsatellite and insertion/deletion markers in a large panel of African populations and found that the Coloured population has equal levels of four ancestries: Southern African Khoisan, Bantu speaking, Indian, and European. The second study conducted by Patterson et al. (2010) genotyped 20 Coloured individuals using genome wide analysis of almost 900,000 markers and concluded that the Coloured population has a substantial genetic contribution from at least four population groups, viz. African (genetically close to isiXhosa), Indonesian, European, and South Asian. Genome wide analysis performed by De Wit et al. (2010) genotyped 959 self-identified Coloured individuals with nearly 500,000 markers using a linkage and admixture model. Their study revealed that the major ancestral contributions were Khoisan (32-43%), Bantu speaking Africans (20-36%), European (21-28%), and to a lesser extent Asian (9-11%), this depending on the model used. The study by Quintana-Murci et al. (2010) analysed mtDNA and NRY lineages of 590 individuals which indicated that the maternal pool was mainly Sub-Saharan African (79%), with a lower frequency for South/South East Asian (16%) and West Eurasian/European lineages (4.6%). The NRY haplogroups revealed that the paternal pool was predominately African (45.2%) than West Eurasian/European (37.72), and with fewer South/South East Asian paternal lineages (17.11%). These findings (De Wit et al., 2010; Quintana-Murci et al., 2010) were particularly interesting as they excluded Cape Muslims from their studies, focusing only on Christian Coloured individuals. The selection criteria for donors in other studies (Tishkoff et al., 2009; Patterson et al., 2010) were not as clearly specified and may have included Cape Muslims, as suggested by De Wit et al. (2010) on commenting the findings of Tishkoff et al. (2009) given the high Asian and European ancestry found in their study. This seems plausible given the results of the present study (Table 1), particularly when one only examines the ancestry of the oldest Cape Muslim subgroups (Cape Coloured Muslims and Cape Malay Muslims). Even though their mtDNA lineages were primarily African, considerable frequencies for Asian and European lineages were also observed. Furthermore, their paternal lineages showed a significant contribution of Asian and European lineages, with only minor African contributions. The studies regarding the ancestry of the Coloured population, particularly the one by Quintana-Murci et al. (2010) indicate that both Cape Muslims and the Coloured population share the same parental populations, even though the mtDNA and NRY distributions of African, Asian and European lineages may vary between these two population groups.
Conclusion
The results of this study showed a clear difference in the contribution of major continent-specific haplogroups to the maternal and paternal lineages of the contemporary Cape Muslim population. Overall admixture estimates for the maternal line indicated Asian (0.4168) and African mtDNA (0.4005) as the main contributors. The admixture estimates for the paternal line however showed a predominance of Asian contribution (0.7852). It should be interesting to further investigate the contribution of other local small communities to the Cape Muslim populations throughout the Western Cape Province of South Africa. The study of the genetic diversity of local communities sharing the same residential areas, such as the Coloured Christian and Muslim communities living in the Kensington-Factreton residential area in the Cape Peninsula, can give genetic evidence of their origins, migration histories, and their relatedness to each other, and especially so the contribution of these communities to each other's gene pool, through intermarriages, conversion and blending.
Acknowledgments
We would like to thank all the donors who participated in the study, as well as everyone who assisted in the collection of samples. We are also grateful to Prof Yusuf Da Costa, Mr. Ebrahim Rhoda, Mr. Ebrahim Salie, Mr. Ebrahim Manual, Prof. Rob Shell, Dr Yassin Mohamed and Dr Mohamed Haron for their valuable contributions. Special thanks to Moulana Igsaan Hendricks, the president of the Muslim Judicial Council of South Africa, for his support for the study. The present study was supported by the National Research Foundation (South Africa).
Internet Resources
Received: November 13, 2012
Accepted: March 12, 2013.
Associate Editor: Francisco M. Salzano
References
- Agrawal S, Khan F, Pandey A, Tripathi M and Herrera RJ (2005) YAP, signature of an African-Middle Eastern migration into northern India. Curr Sci 8:1977-1980.
- Agresti, A (1990) Categorical Data Analysis. Wiley Interscience, New York, 558 pp.
- Alves-Silva J, da Silva Santos M, Guimaraes PEM, Ferreira ACS, Bandelt HJ, Pena SDJ and Prado VF (2000) The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67:444-461.
- Alzualde, A, Izagirre N, Alonso S, Alonso A and De La Rúa C (2005) Temporal mitochondrial DNA variation in the Basque country: Influence of post-Neolithic events. Annu Hum Genet 69:665-679.
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH and Wallace DC (1992) Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient Mongoloid migrations. Genetics 130:139-152.
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, et al. (2003) Ethnic India: A genomic view, with special reference to peopling and structure. Genome Res 13:2277-2290.
- Beleza S, Gusmão L, Lopes A, Alves C, Gomes I, Giouzeli M, Calafell F, Carracedo A and Amorim A (2005) Microphylogeographic and demographic history of Portuguese male lineages. Ann Hum Genet 70:181-194.
- Bermisheva MA, Tamberts K, Villems R and Khusnutdinova EK (2002) Diversity of mitochondrial DNA haplogroups in ethnic populations of the Volga-Ural region. Mol Biol 36:802-812.
- Berniell-Lee G, Plaza S, Bosch E, Calafell F, Jourdan E, Césari M, Lefranc G and Comas D (2008) Admixture and sexual bias in the population settlement of La Réunion Island (Indian Ocean). Am J Physical Anthropol 136:100-107.
- Bickford-Smith V (1994) Meanings of freedom: Social position and identity among ex-slaves and their decendants in Cape Town. In: Worden N and Crais C (eds) Breaking the chains: Slavery and its legacy in the 19th century Cape Colony. Witwatersrand University Press, Johannesburg, pp 289-312.
- Bradlow FR and Cairns M (1978) The early Cape Muslims: A study of their mosques, genealogy and origins. Printpak, Cape Town, pp 80-99.
- Byrne EM, McRae AF, Zhao ZZ, Martin NG, Montgomery GW and Visscher PM (2008) The use of common mitochondrial variants to detect and characterise population structure in the Australian population: Implications for genome-wide association studies. Eur J Hum Genet 16:1396-1403.
- Cavalli-Sforza LL and Feldman MW (2003) The application of molecular genetic approaches to the study of human evolution. Nat Genet 33:266-274.
- Chakraborty R (1985) Gene identity in racial hybrids and estimation of admixture rates. In: Neel JV and Ahuja Y (eds) Genetic Microdifferentiation in Man and Other Animals. Indian Anthropological Association, Delhi, pp 171-180.
- Chen Y, Olckers A, Schurr T, Kogelnik AM, Huoponen K and Wallace, DC (2000) mtDNA variation in the South African Kung and Khwe -And their genetic relationships to other African populations. Am J Hum Genet 66:1362-1383.
- Cloete K, Ehrenreich L, D'Amato ME, Leat N, Davison S and Benjeddou M (2010) Analysis of seventeen Y-chromosome STR loci in the Cape Muslim population of South Africa. Legal Med 12:42-45.
- Cordaux R, Saha N, Bentley GR, Aunger R, Sirajuddin SM and Stoneking M (2003) Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet 11:253-264.
- Cordaux R, Weiss G, Saha N and Stoneking M (2004) The Northeast Indian passageway: A barrier or corridor for human migrations. Mol Biol Evol 21:1525-1533.
- Cruciani F, Santolamazza P, Shen P, Macualay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, et al. (2002) A back migration from Asia to Sub-Saharan Africa is supported by high-Resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:11971214.
- Da Costa Y (1994) Muslims in Greater Cape Town: A problem of identity. Brit J Sci 45:236-246.
- Da Costa Y and Davids A (1994) Pages from Cape Muslim History. Shuter and Shooter, Pietermaritzburg, 182 pp.
- Davids A (1980) The mosques of the Bo-Kaap: A Social History of Islam at the Cape. Cape and Transvaal Printers, Parow, 236 pp.
- Debnath M, Palanichamy MG, Mitra B, Jin JQ, Chaudhuri TK and Zhang YP (2011) Y chromosome diversity in the sub-Himalayan Terai and Duars population of East India. J Hum Genet 56:765-771.
- De Wit E, Delport W, Rugamika CE, Meintjies A, Möller M, van Helden PD, Seoighe C and Hoal EG (2010) Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape. Hum Genet 128:145-153.
- Du Pre RH (1994) Separate but unequal. The Coloured' People of South Africa -A Political History. Jonathan Ball Publishers, Johannesburg, 11 pp.
- Edwin D, Vishwanathan H, Roy S, Usha Rani MV and Mujumder PP (2002) Mitochondrial DNA diversity among five tribal populations of southern India. Curr Sci 83:158-162.
- Flores C, Maca-Meyer N, Pérez JA, González AM, Larruga JM and Cabrera VM (2003) A predominant European ancestry of paternal lineages from Canary islanders. Ann Hum Genet 67:138-152.
- Fregel R, Gomes V, Gusmão L, González1 AM, Cabrera1 VM, Amorim A and Larruga JM (2009) Demographic history of Canary Islands male gene-pool: replacement of native lineages by European. BMC Evol Biol 9:181.
- Giliomee H (2003) The Afrikaners: Biography of a People. Tafelberg, Cape Town, 90 pp.
- Gonder ML, Mortensen MK, Reed FA, de Sousa A and Tishkoff SA (2007) Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol 24:757-768.
- Gauniyal, M, Chahal SMS and Kshatriya GK (2008) Genetic affinities of the Siddis of South India: An emigrant population of East Africa. Hum Biol 80:251-270.
- Hammer MF and Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951-962.
- Hammer MF and Zegura SL (2002) The human Y chromosome haplogroup tree: Nomenclature and phylogeography of its major divisions. Annu Rev Anthropol 31:303-321.
- Hurles ME, Sykes BC, Jobling MA and Forster P (2005) The dual origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from maternal and paternal lineages. Am J Hum Genet 76:894-901.
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL and Hammer MF (2008) New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18:830-838.
- Kayser M, Brauer S, Weiss G, Schiefenhövel W, Underhill P, Shen P, Oefner P, Tommaseo-Ponzetta M and Stoneking M (2003) Reduced Y-chromosome, but mitochondrial DNA diversity in human populations from West New Guinea. Am J Hum Genet 72:281-302.
- Kayser M, Lao O, Anslinger K, Augustin C, Bargel G, Edelmann J, Elias S, Heinrich M, Henke J, Hohoff C, et al. (2005) Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum Genet 117:428-443.
- Keegan T (1996) Colonial South Africa and the Origins of the Racial Order. University of Virginia Press, Charlottesville, 368 pp.
- Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S and Villems R (1999) The place of the Indian mtDNA variants in the global network of maternal lineages and the peopling of the Old World. In: Papiha SS, Deka R and Chakraboty R (eds) Genomic Diversity: Applications in Human Population Genetics. Springer, New York, pp 135-152.
- Krithika S, Maji S and Vasulu TS (2007) Molecular genetic perspective of Indian populations: A Y-chromosome scenario. Anthropol Special 3:385-392.
- Kumar V, Reddy ANS, Babu JP, Tipirisetti NR, Langstieh BT, Thangaraj K, Reddy AG, Singh L and Reddy BM (2007) Y-chromosome evidence suggests a common heritage of Austro-Asiatic populations. BMC Evol Biol 7:47.
- Lell JT and Wallace DC (2000) The peopling of Europe from the maternal and paternal Perspectives. Am J Hum Genet 67:1376-1381.
- Lodhi AY (1992) African settlements in India. Nordi J Afr Stud 1:83-86.
- Loogväli EL, Roostalu U, Malyarchuk BA, Derenko MV, Kivisild T, Metspalu E, Tambets K, Reidla M, Tolk HV, Parik J et al. (2004) Disuniting uniformity: A pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol 21:2012-2021.
- Mahida EM (1993) History of Muslims in South Africa: A Chronology. Kat Bros, Durban, 154 pp.
- Maji S, Krithika S and Vasulu TS (2009) Phylogeographic distribution of mitochondrial DNA macrohaplogroup M in India. J Genet 88:127-139.
- Manwaring N, Jones MM, Wang JJ, Rochtchina E, Mitchell P and Sue CM (2006) Prevalence of mitochondrial DNA haplogroup in an Australian population. Int Med J 36:530-533.
- Martinez-Cruzado JC, Toro-Labrador G, Ho-Fung V, Estévez-Montero MA, Lobaina-Manzanet A, Padovani-Claudio DA, Sánchez-Cruz H, Ortiz-Bermúdez P and Sánchez-Crespo A (2001) Mitochondrial DNA analysis reveals substantial Native American ancestry in Puerto Rico. Hum Biol 73:491-511.
- Merriwether DA, Friedlaender JS, Mediavilla J, Mgone C, Gentz F, Ferrell RE (1999) Mitochondrial DNA variation is an indicator of Austronesian influence in Island Melanesia. Am J Phys Anthropol 110:243-270.
- Mishmar D, Ruis-Pesini E, Golik P, Macualay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171-176.
- Mountain A (2003) The First People of the Cape. David Philips Publishers, Cape Town, 102 pp.
- Patterson N, Petersen DC, Van der Ross RE, Sudoyo H, Glashoff RH, Marzuki S, Reich D and Hayes VM (2010) Genetic structure of a unique admixed population: Implication for medical research. Hum Mol Genet 19:411-419.
- Pickel B (1997) Coloured Ethnicity and Identity: A Case Study in the Former Coloured Areas of the Western Cape South Africa. Demokratie und Entwicklung Series No. 28. Hamburg, 124 pp.
- Piechota J, Tonska K, Nowak M, Kabzinska D, Lorenc A, Bartnik E (2004) Comparison between the Polish population and European populations on the basis of mitochondrial morphs and haplogroups. Acta Biochim Polon 51:883-895.
- Quintana-Murci L, Harmant C, Quach H, Balanovsky O, Zaporozhchenko V, Bormans C, Van Helden PD, Hoal EG and Behar DM (2010) Strong maternal Khoisan contribution to the South African Coloured population: A case of gender-biased admixture. Am J Hum Genet 86:611-620.
- Rootsi S, Magri C, Kivisild T, Benuzzi G, Help H, Bermisheva M, Kutuev I, Barac L, Pericsic M, Balanovsky O, et al. (2004) Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am J Hum Genet 75:128-137.
- Roychoudhury S, Roy S, Badal D, Chakraborty M, Roy M, Roy B, Ramesh A, Prabhakaran, N, Usha Rani MV, Vishwanathan H, et al. (2000) Fundamental genomic unity of ethnic India is revealed by analysis of mitochondrial DNA. Curr Sci 79:1182-1192.
- Santos C, Montiel R, Angles N, Lima M, Francalacci P, Malgosa A, Abade A and Aluja MP (2004) Determination of human Caucasian mitochondrial DNA by means of a hierarchical approach. Hum Biol 76:431-453.
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, Benedictis GD, Francalacci P, Kouvatsi A, Limborska S, et al. (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome Perspective. Science 290:1155-1159.
- Shah AM, Tamang R, Moorjani P, Rani DS, Govindaraj P, Kulkarni G, Bhattacharya T, Mustak MS, Bhaskar LV, Reddy AG et al. (2011) Indian Siddis: African descendants with Indian Admixture. Am J Hum Genet 89:154-61.
- Schurr TG and Wallace DC (2002) Mitochondrial DNA diversity in Southeast Asian populations. Hum Biol 74:431-452.
- Shell RCH (2000) Islam in Southern Africa. In: Levtzion N and Pouwels L (eds) The History of Islam in Africa. Ohio University Press, Athens, pp 327-348.
- Stone AC and Stoneking M (1998) mtDNA analysis of a prehistoric Oneata population: Implications for the peopling of the New World. Am J Hum Genet 62:1153-1170.
- Sudoyo H, Suryadi H, Pramoonjago PLP, Lyrawati D and Marzuki S (2002) Asian-specific mtDNA backgrounds associated with the primary G11778A mutation of Leber's hereditary optic neuropathy. J Hum Genet 47:594-604.
- Tajima A, Hayami M, Juji KTK, Matsuo M, Marzuki S, Omoto K and Horai S (2004) Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages. J Hum Genet 49:187-193.
- Tishkoff SA, Reed FA, Friendlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. (2009) The genetic structure and history of Africans and African Americans. Science 324:1035-1044.
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus M and Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetetics 144:1835-1850.
- Torroni A, Achilli A, Macaualay V, Richards M and Bandelt HJ (2006) Harvesting the fruit of the human mtDNA tree. Trends Genet 22:340-345
- Tripathy V, Nirmala A and Reddy BM (2008) Trends in molecular anthropological studies in India. Int J Hum Genet 8:1-20.
- Underhill PA (2004) A synopsis of extant Y chromosome diversity in East Asia and Oceania. In: Sagart L, Roger M, Blench RM and Sanchez-Mazas A (eds) The peopling of East Asia: Putting Together Archaeology, Linguistics and Genetics. Routledge Curzon, London, pp 301-319.
- Underhill PA and Kivisild T (2007) Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet 41:539-64.
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL and Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996-1005.
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, et al. (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358-361.
- Underhill PA, Passarino G, Lin AA, Shen P, Lahr MM, Foley RA, Oefner PJ and Cavalli-Sforza LL (2001) The phyleogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43-62.
- Van Oven M and Kayser M (2008) Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Mutat Brief 29:E386-E394.
- Wallace DC, Brown MD and Lott MT (1999) Mitochondrial DNA variation in human evolution and disease. Gene 238:211-230.
- Wells S (2007) Deep Ancestry: Inside the Genographic Project. National Geographic, Washington D.C., 247 pp.
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339-348.
-
ADMIX 95 Software, http://www.genetica.fmed.edu.uy/software.htm (February 17, 2013).
» link
Publication Dates
-
Publication in this collection
19 Apr 2013 -
Date of issue
2013
History
-
Received
13 Nov 2012 -
Accepted
12 Mar 2013

 Reconstruction of major maternal and paternal lineages of the Cape Muslim population
Reconstruction of major maternal and paternal lineages of the Cape Muslim population