Abstract
Seven isolates from patients with American cutaneous leishmaniasis in the Amazon region of Brazil were phenotypically suggestive of Leishmania (Viannia) guyanensis/L. (V.) shawi hybrids. In this work, two molecular targets were employed to check the hybrid identity of the putative hybrids. Heat shock protein 70 (hsp70) gene sequences were analyzed by three different polymerase chain reaction (PCR) approaches, and two different patterns of inherited hsp70 alleles were found. Three isolates presented heterozygous L. (V.) guyanensis/L. (V.) shawi patterns, and four presented homozygous hsp70 patterns involving only L. (V.) shawi alleles. The amplicon sequences confirmed the RFLP patterns. The high-resolution melting method detected variant heterozygous and homozygous profiles. Single-nucleotide polymorphism genotyping/cleaved amplified polymorphic site analysis suggested a higher contribution from L. (V.) guyanensis in hsp70 heterozygous hybrids. Additionally, PCR-RFLP analysis targeting the enzyme mannose phosphate isomerase (mpi) gene indicated heterozygous and homozygous cleavage patterns for L. (V.) shawi and L. (V.) guyanensis, corroborating the hsp70 findings. In this communication, we present molecular findings based on partial informative regions of the coding sequences of hsp70 and mpi as markers confirming that some of the parasite strains from the Brazilian Amazon region are indeed hybrids between L. (V.) guyanensis and L. (V.) shawi.
Keywords:
Cutaneous Leishmaniaisis; hybrid parasite; L. (V.) guyanensis; L. (V.) shawi; Brazil
Introduction
The genus Leishmania presents highly diverse nonsexual mechanisms for the generation of diversity. These mechanisms include the occurrence of tandem repeat genes, gene amplification, gene duplication, mini-chromosome generation and mosaic aneuploidy (Delgado et al., 1997Delgado O, Cupolillo E, Bonfante-Garrido R, Silva S, Belfort E, Grimaldi G Júnior and Momen H (1997) Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem Inst Oswaldo Cruz 92:581-582.; Victoir and Dujardin, 2002Victoir K and Dujardin JC (2002) How to succeed in parasitic life without sex? Asking Leishmania. Trends Parasitol 18:81-85.; Dujardin et al., 2007Dujardin JC, De Doncker S, Jacquet D, Bañuls AL, Balavoine M, van Bockstaele D, Tibayrenc M, Arevalo J and Le Ray D (2007) Clonal propagation and the fast generation of karyotype diversity: an in vitro Leishmania model. Parasitology 134:33-39.; Sterkers et al., 2011Sterkers Y, Lachaud L, Crobu L, Bastien P and Pagès M (2011) FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol 13:274-283.). This high genome plasticity is achieved through both sexual-like and nonsexual characteristics, providing great complexity that is reflected in the wide geographical distribution of Leishmania spp., the diversity of its hosts and the complexity of illnesses associated with the parasites.
New tools for the study of the reproduction mechanisms of the Leishmania genus have expanded the knowledge of these mechanisms, showing that parasite population structure is predominantly clonal, but rare sexual events can occur (Tibayrenc and Ayala, 1991Tibayrenc M and Ayala FJ (1991) Towards a population genetics of microorganisms: the clonal theory of parasitic protozoa. Parasitol Today 7:228-232.; Tibayrenc et al., 1991Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Brenière SF, Dardé ML and Ayala FJ (1991) Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci U S A 88:5129-5133.). Nevertheless, genetic exchange between Leishmania parasites as well as other related trypanosomatids has been experimentally demonstrated, and complex analyses have been performed in hybrid strains (Gaunt et al., 2003Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, Taylor MC, Mena SS, Veazey P, Miles GAJ, Acosta N, et al. (2003) Mechanism of genetic exchange in american trypanosomes. Nature 421:936-939.; Akopyants et al., 2009Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM and Sacks DL (2009) Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324:265-268.; Rogers et al., 2014Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA and Smith DF (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092.; Inbar et al., 2019Inbar E, Shaik J, Iantorno SA, Romano A, Nzelu CO, Sanders MJ, Dobson D, Cotton JA, Grigg ME, Beverley SM, et al. (2019) Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania PLoS Genet 15:e1008042.).
Reports of hybrids occurring in natural environments or involved in cases of leishmaniasis are recurring worldwide. In some cases, these hybrids are associated with severe forms of the disease, including the mucosal form found in Peru (Dujardin et al., 1995Dujardin JC, Bañuls AL , Llanos-Cuentas A, Alvarez E, DeDoncker S, Jacquet D, Le Ray D, Arevalo J and Tibayrenc M (1995) Putative Leishmania hybrids in the eastern Andean valley of Huanuco, Peru. Acta Trop 59:293-307.). The occurrence of hybrids has been directly correlated with areas of sympatric species occurrence, which enables the interaction of various genotypes in coinfected vectors (Inbar et al., 2013Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, Kauffmann F, Barhoumi M, Grigg M, Owens K, et al. (2013) The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672.; Kato et al., 2019Kato H, Gomez EA, Seki C, Furumoto H, Martini-Hobles L, Muzzio J, Calvopiña M, Velez L, Kubo M, Tabbabi A, et al. (2019) PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. Plos Negl Trop Dis 13:e0007403.). Studies have shown that significant inbreeding of parasites from strains that are genetically related or even identical can occur in the vector host (Rougeron et al., 2009Rougeron V, Meeûs TD, Hide M, Waleckx E, Bermudez H, Arevalo J, Llanos-Cuentas A, Dujardin JC, De Doncker S, Le Ray D, et al. (2009) Extreme inbreeding in Leishmania braziliensis. PNAS 106:10224-10229.). The low frequency of coinfection in the invertebrate host represents the major barrier to mating between unrelated strains (Bastien et al., 1992Bastien P, Blaineau C and Pages M (1992) Leishmania: sex, lies and karyotype. Parasitol Today 8:174-177.; Inbar et al., 2013Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, Kauffmann F, Barhoumi M, Grigg M, Owens K, et al. (2013) The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672.). Recent studies have shown that intraspecies hybrids are prone to hybridization events, while interspecies hybrids seem to be sterile (Inbar, et al., 2019Inbar E, Shaik J, Iantorno SA, Romano A, Nzelu CO, Sanders MJ, Dobson D, Cotton JA, Grigg ME, Beverley SM, et al. (2019) Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania PLoS Genet 15:e1008042.).
In the lower Amazon region of Pará State, Brazil, reports indicate the circulation of at least five different species of Leishmania, involved with American cutaneous leishmaniasis (ACL) cases, in addition to two different subpopulations of L. (V.) shawi, classified as L. (V.) shawi shawi and L. (V.) shawi santarensis (Jennings et al., 2014Jennings YL, Souza AAA, Ishikawa EA, Shaw JJ, Lainson R and Silveira FT (2014) Phenotypic characterization of Leishmania spp causing cutaneous leishmaniasis in the lower Amazon region, Western Pará State, Brazil, reveals a putative hybrid parasite, Leishmania (Viannia) guyanensis×Leishmania (Viannia) shawi shawi. Parasite 21:39.). The authors also isolated seven atypical strains from human cases of ACL from Santarém, a city located in the lower Amazon region. By means of multilocus enzyme electrophoresis (MLEE) and monoclonal antibody analysis, these atypical strains presented phenotypic profiles suggestive of hybridization between the species L. (V.) guyanensis and L. (V.) shawi (Table S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. ).
The verification of the L. (V.) guyanensis/ L. (V.) shawi hybrid parasites involved in ACL cases in the Santarém region updates the eco-epidemiological scenario in this important endemic area of Brazil. Therefore, the aim of the present study was to search for evidence of genetic recombination in these strains using hsp70 sequences as targets for polymerase chain reaction and restriction fragment length polymorphisms (PCR-RFLP), single-nucleotide polymorphism genotyping/cleaved amplified polymorphic site (SNP-CAPS), high-resolution melting PCR (HRM) and PCR product sequencing analyses. An additional PCR-RFLP analysis of mpi sequences for comparison with hsp70 findings was performed. Together, our findings confirmed by molecular methods that five of seven isolates presented genetic traces of hybrid strains between L. (V.) shawi and L. (V.) guyanensis.
Material and Methods
Parasites
The Leishmania strains were obtained from the Evandro Chagas Institute (Surveillance Secretary of Health, Ministry of Health), Pará State, Brazil and are listed in Table 1. These strains included 21 reference strains of the main species present in the lower Amazon region, including four different reference strains of L. (V.) guyanensis, two of L. (V.) shawi shawi, two of L. (V.) shawi santarensis and the seven putative L. guyanensis /L. shawi hybrids that were isolated from the skin lesions of patients with localized cutaneous leishmaniasis.
Cloning of the parasites
Two different methodologies were used for Leishmania spp. cloning.
The cloning protocol involved plating the parasites in solid growth media according to Muniaraj et al. (2010Muniaraj M, Sinha PK and Das P (2010) Antileishmanial activity of drug infused mini-agar plates on Leishmania donovani promastigotes. Trop Biomed 27:657-661. ) with some modifications. We chose to prepare the culture plates using 0.5 volumes (V) of 2x Noble agar medium (Sigma; St. Louis, MO, USA) and 0.5 V of 2x Schneider medium (Sigma; St. Louis, MO, USA), with a final fetal bovine serum (FBS) concentration of 10%. Then, 103 cells were seeded per plate, followed by incubated at 25 °C for 30 days.
The second cloning method was performed according to Handman et al. (1983Handman E, Hocking RE, Mitchell GF and Spithill TW (1983) Isolation an characterization of infective and non-infective clones of Leishmania tropica. Mol Biochem Parasitol 7:111-126. ) with modifications that are briefly described here. The parasites were cultivated in Schneider´s insect medium with 10% FBS, and stationary-phase cultures were cloned via limiting dilution assays at concentrations from 6.2 x 104 to 0.03 parasites/mL. Cultures were expanded from wells containing approximately one parasite per mL.
DNA extraction
DNA was obtained from each culture by phenol-chloroform extraction followed by precipitation with sodium acetate and ethanol (Uliana et al., 1991Uliana SR, Affonso MH, Camargo EP and Floeter-Winter LM (1991) Leishmania: genus identification based on a specific sequence of the 18S ribosomal RNA sequence. Exp Parasitol 72:157-163.), then resuspended in Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.4).
DNA and PCR product quantification
The purified PCR products and DNA obtained from the parasite cultures were quantified using a BioSpectrometer (Eppendorf) according to the manufacturer’s instructions.
hsp70 PCR-RFLP analysis
DNA from five clones from each isolate strain and the reference strains was used in the PCR assays. Each PCR assay was performed in a final volume of 50 µL containing 50 ng of DNA, each primer at 0.2 µM and 25 µL of Top Taq Master Mix (Qiagen, Germantown, USA). The primers used for hsp70 were hsp70c forward, 5’-GGACGAGATCGAGCGCATGGT-3’, and hsp70c reverse, 5’-TCCTTCGACGCCTCCTGGTTG-3’, which amplify a 234 base pair (bp) fragment (Graça et al., 2012Graça GC, Volpini AC, Romero GAS, Oliveira Neto MP, Hueb M, Porrozzi R, Boité MC and Cupolillo E (2012) Development and validation of PCR-based assays for diagnosis of american cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz 107:664-674.). The PCR assays were conducted with the following amplification program: 94 °C for 5 min, by 40 cycles of 94 °C for 1 min, 64 °C for 1 min and 72 °C for 30 s, with a final extension at 72 °C for 10 min. The reaction product was analyzed by electrophoresis on a 2% agarose gel in TAE buffer (40 mM Tris-acetate and 2 mM EDTA). DNA fragments were stained with Gel Red (Biotium, Freemont, CA, USA) and were visualized in a transilluminator Gel Logic 212 Pro® system (Carestream Molecular Imaging; Woodbridge, CT, U.S.A) at 260 nm. Images were captured with a Pro imaging GL212 camera using an orange filter.
The hsp70 PCR-RFLP analysis for discrimination between L. (V.) guyanensis and L. (V.) shawi was performed as described by Graça et al. (2012Graça GC, Volpini AC, Romero GAS, Oliveira Neto MP, Hueb M, Porrozzi R, Boité MC and Cupolillo E (2012) Development and validation of PCR-based assays for diagnosis of american cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz 107:664-674.). The PCR products were purified using a GeneJet PCR purification Kit (Thermo Fisher Scientific, Waltham, MA, USA), and 300 ng of purified PCR product was digested with 10 units (U) of HaeIII (Thermo Scientific-Walthman, MA, USA) using the conditions recommended by the manufacturer. Incubation was performed at 37 °C for 12 hours. The digested products were subjected to 3% agarose gel electrophoresis and stained with GelRed. The visualization of fragments was performed as described above.
mpi PCR-RFLP analysis
The mpi PCR-RFLP analysis for discrimination between L. (V.) guyanensis and L. (V.) shawi was performed as described by Boité et al. (2012Boité MC, Mauricio IL, Miles MA and Cupolillo E (2012) New insights on taxonomy, phylogeny and population genetics of Leishmania (Viannia) parasites based on multilocus sequence analysis. Plos Negl Trop Dis 6:e1888. ). DNA from five clones from each isolate strain and the reference strains were used for PCR. Each amplification reaction was performed in a final volume of 50 µL containing 50 ng of DNA, each primer at 2 µM and 25 µL of Top Taq Master Mix (Qiagen, Germantown, USA). The primers used were mpi forward, 5’- GGCAAGATGTATGCGGAGTT-3’, and mpi reverse 5’-TCCTTCGACGCCTCCTGGTTG-3’, which amplify a 681 bp fragment. The PCR assays were performed with the following amplification program: 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 58 °C for 1 min and 72 °C for 30 s, with by a final extension at 72 °C for 10 min. The reaction product was analyzed by electrophoresis on a 2% agarose gel in TAE buffer (40 mM Tris-acetate and 2 mM EDTA). DNA fragments were stained with Gel-Red and were visualized in a transilluminator Gel Logic 212 Pro® system at 260 nm. Images were captured with a Pro imaging GL212 camera using an orange filter.
The PCR products were purified using a GeneJet PCR purification Kit (Thermo Fisher Scientific, Waltham, MA, USA), and 300 ng of the purified PCR product was digested with 1 U of Cla I (New England BioLabs- Ipswich, MA, EUA) under the conditions recommended by the manufacturer. Incubation was performed at 37 °C for 2 h. The enzyme was heat inactivated at 65 °C for 15 min. The digested products were subjected to 2% agarose gel electrophoresis and stained with GelRed. The visualization of fragments was performed as described above.
Cloning and sequencing of hsp70 PCR products
The hsp70c PCR products were ligated into the PGEM T Easy vector (PROMEGA-Madison, Wisconsin, EUA) and cloned into E. coli bacteria (SURE). The hsp70 amplicons were sequenced by Sanger sequencing (Big Dye Terminator V3.1 cycle sequencing kits; Applied Biosystems). PCR product sequencing was performed for five putative Leishmania hybrid isolates (isolates 2 to 6) as well as two strains of L. (V.) guyanensis and L. (V.) shawi. Three independent amplification reactions were performed for each isolate. Six plasmids containing PCR product clones were chosen from each PCR assay, from which four PCR product clones were sequenced.
SNP-CAPS analysis of putative hybrids and progeny lines
The proportion of each hsp70 allele in each isolate was determined by comparing the relative intensity of the PCR products between putative hybrids and progeny lines after SNP-CAPS analysis (Akopyants et al., 2009Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM and Sacks DL (2009) Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324:265-268.). The primers and PCR conditions were the same as those indicated above. A total of 300 ng of the purified hsp70 amplicon from the putative hybrid or parental reference strain was cleaved using 10 U of HaeIII at 37 °C for 12 h. The intensity of the bands was compared to three amplicon mixtures of 300 ng of the purified hsp70 PCR products from the DNA of the parental strains L. (V.) guyanensis and L. (V.) shawi at proportions of 2:1, 1:1 and 1:2, respectively. An analysis of the relative intensity of bands was performed using Carestream Molecular Imaging Software.
HRM analyses
The hybrids isolates were evaluated via HRM analyses to observe possible discriminatory patterns capable of indicating allelic variations. Real-time PCR assays were performed as described by Zampieri et al. (2016Zampieri RA, Laranjeira-Silva MF, Muxel SM, Lima ACS, Shaw JJ and Floeter-Winter LM (2016) High Resolution Melting Analysis targeting hsp70 as a fast and efficient method for the discrimination of Leishmania species. PLoS Negl Trop Dis 10:e0004485. ) in PikoReal96 thermocycler (Thermo Fisher Scientific, Waltham, MA, USA) using MeltDoctor HRM Master Mix (Life Technologies; Carlsbad, California, USA) with 50 ng of genomic DNA as the template and the hsp70F2 (5’-GGAGAACTACGCGTACTCGATGAAG-3’) and hsp70c reverse primers at 200 nM, as described above. The cycling conditions were 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s and annealing/extension at 60 °C for 30 s, with fluorescent signal acquisition at the end of each extension step, followed by the dissociation curve for HRM analysis. The amplicon dissociation analyses were performed via the acquisition of fluorescence signals at 0.2 °C intervals with holding for 10 s between 60 °C and 95 °C. Genomic DNA samples from the reference strains L. (L.) infantum chagasi (MCER/BR/1981/M6445), L. (L.) amazonensis (MHOM/BR/1973/M2269), L. (L.) mexicana (MNYC/BZ/62/M379), L. (L.) lainsoni (MHOM/BR/81/M6426), L. (V.) braziliensis (MHOM/BR/1975/M2903), L. (V.) guyanensis (MHOM/BR/1975/M4147), L. (V.) naiffi (MDAS/BR/1979/M5533) and L. (V.) shawi (MCEB/BR/84/M8408) were used as standards, and the HRM profiles generated for these species were the benchmark of the analysis.
Results
Homozygous or heterozygous RFLP profiles were detected in hybrid strains
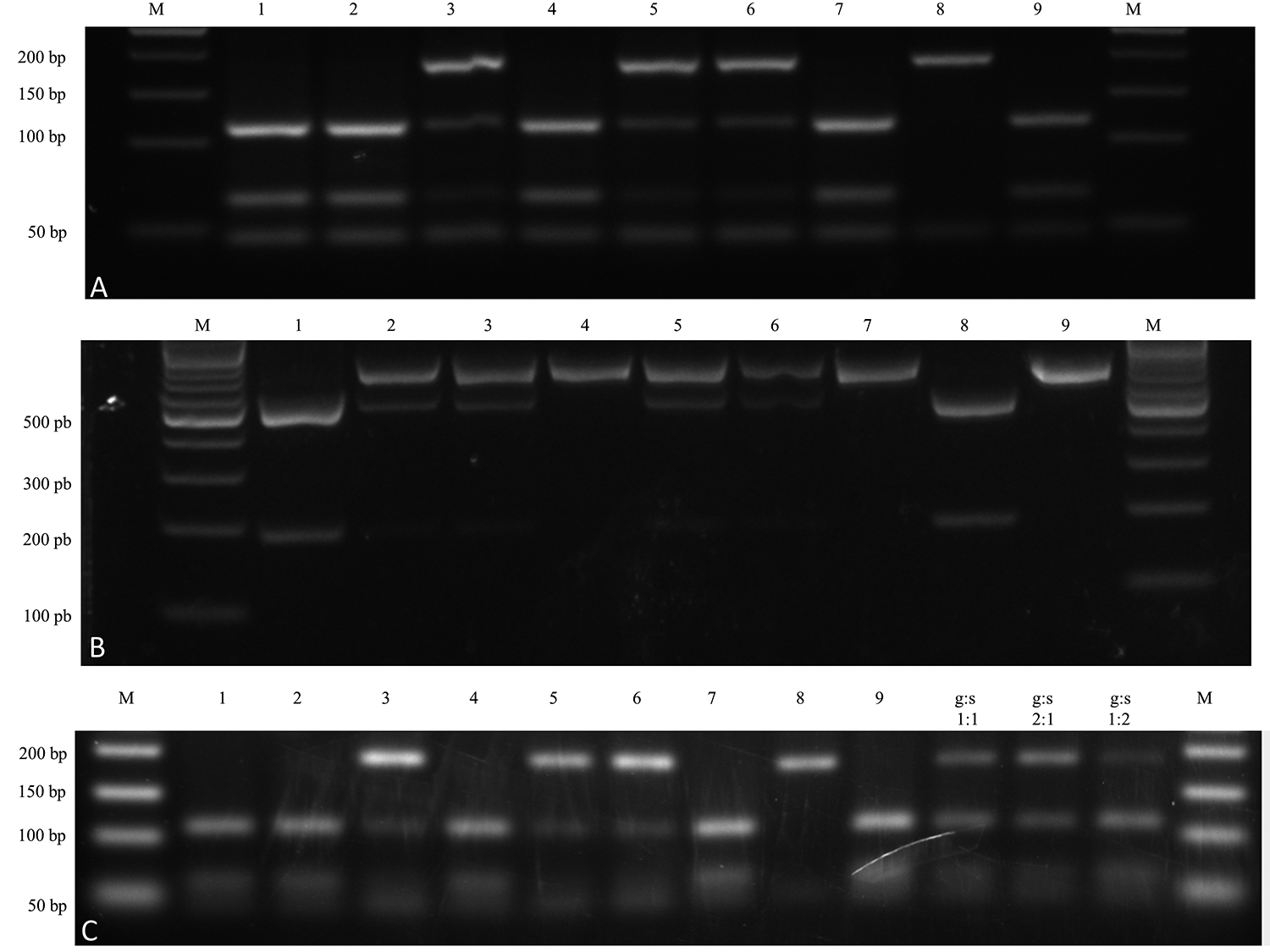
The analysis of the results of the HaeIII digestion of the hsp70 PCR products and the Cla I digestion of the mpi PCR products was performed for seven isolates (Figure 1A and Figure 1B), as well as for five clones obtained from each isolate (Figure S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. ). It was shown, that the hybrid phenotype of the isolates was not due to mixed infection, since all the clones from each isolate presented the same cleavage profiles in RFLP assays (Figure S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. ).
Electrophoresis gel fractionated products on agarose stained with GelRed showing homozygous and heterozygous PCR-RFLP patters. A: hsp70 PCR-RFLP (HaeIII). M: 50 bp molecular weight marker (Fermentas SM 0373). Lanes 1 to 7: isolates 1 to 7, respectively. Lane 8: L. (V.) guyanensis (MHOM/BR/1975/M4147). Lane 9: L. (V.) shaw shawi (MCEB/BR/1984/M8408). B: mpi PCR-RFLP (Cla I). M: 100 bp molecular weight marker (Fermentas SM 0331). Lanes 1 to 7: isolates 1 to 7, respectively. Lane 8: L. (V.) guyanensis (MHOM/BR/1975/M4147). Lane 9: L. (V.) shaw shawi (MCEB/BR/1984/M8408). C: SNP-CAPS hsp70 PCR-RFLP (HaeIII). All lanes contain 300 ng of purified PCR products digested by HaeIII. Lanes 1 to 7: hybrids Leishmania (Viannia) spp. isolates 1 to 7, respectively. Lane 8: L. (V.) guyanensis; Lane 9: L. (V.) shawi shawi; Lane 10: L. (V.) guyanensis: L. (V.) shawi shawi (1:1); Lane 11: L. (V.) guyanensis: L. (V.) shawi shawi (2:1); Lane 12: L. (V.) guyanensis: L. (V.) shawi shawi (1:2); and M: 50 bp molecular weight marker.
HaeIII cleavage of the PCR products generated three fragments of 47, 67 and 120 base pairs for L. (V). shawi and two fragments of 47 and 187 base pairs for L. (V.) guyanensis as shown by the hsp70-RFLP analysis of these putative parental strains. Four different strains of these species from the same geographic area were included to assure more consistent results (Figure S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. ). The hsp70 PCR products obtained from the seven isolates generated two RFLP profiles, one that was characteristic of heterozygous alleles, while the other was characteristic of homozygous profile of the L. (V). shawi alleles (Figure 1A).
Three isolates (isolates 3, 5 and 6) showed a heterozygous profile resembling the pattern for L. (V.) shawi and L. (V.) guyanensis with fragments characteristic of both species (Figure 1A). The isolates 1, 2, 4 and 7 presented the hsp70 pattern that was compatible with the homozygous profile of the L. (V). shawi alleles (Figure 1A). To verify these polymorphic profiles in more detail, the hsp70c PCR products were cloned and sequenced. The restriction sites of the HaeIII enzyme in the sequences corresponded to the cleavage pattern obtained in the RFLP assay (Figure S2 Table S2 - NCBI sequences accession numbers. ). Five isolates were chosen for sequencing: three from the heterozygous hsp70 profile and two from the homozygous hsp70 isolates. The hsp70 PCR nucleotide sequences were deposited in GenBank with the accession numbers MT337389 to MT337400 (Table S2 Table S2 - NCBI sequences accession numbers. ). The nucleotide sequences of the hsp70 PCR products obtained from the heterozygous hsp70 strains presented profiles corresponding to L. (V.) shawi and L. (V.) guyanensis, as could be observed from the cloned amplicons of the same sample (Figure S2). The presence of both alleles confirmed the PCR-RFLP pattern presented by the heterozygous isolates. The homozygous cloned PCR nucleotide sequences presented only L. (V) shawi alleles.
Cla I cleavage did not cut the mpi PCR product of L. (V.) shawi and generated two fragments for L. (V.) guyanensis of 184 and 497 base pairs. Two of seven isolates (isolates 4 and 7) presented homozygous profiles compatible with L. (V.) shawi. One isolate (isolate 1) presented a homozygous profile compatible with L. (V.) guyanensis. Four of seven isolates (isolates 2, 3, 5 and 6) presented a heterozygous hybrid pattern of mpi alleles (Figure 1B). Altogether, the hsp70 and mpi PCR-RFLP data reinforced the phenotypic findings favoring the hybrid identity for five of seven (5/7) of these isolates (Figure 1, Figure S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. and Table 2). The seven isolates were divided into four groups when the molecular targets were analyzed together.
hsp70 SNP-CAPS analysis of the putative hybrids
To perform a qualitative analysis of the proportion of parental loci in the hsp70 inheritance profile of the hsp70 hybrid strains, SNP-CAPS assays were performed, allowing a comparative analysis of the intensity of the generated fragments with the parental and uncloned hybrid strain profiles.
The putative hsp70 homozygous hybrids presented bands with intensity similar to that in L. (V.) shawi, suggesting an equivalent number of hsp70 copies. The putative hsp70 heterozygous showed fragments corresponding to L. (V.) guyanensis with higher intensities (Figure 1C). The analysis of the relative intensity of the bands was performed using Carestream Molecular Imaging Software in relation to the heterozygotes, showing that the L. (V.) guyanensis bands were approximately six times brighter those of the heterozygous isolates (Figure S3 Figure S1 - Illustrative gel showing hsp70 and mpi PCR-RFLP (HaeIII) products from cloned parasites fractionated on 3% agarose stained with GelRed. ).These results suggest that L. (V.) guyanensis may have made a greater contribution to the inheritance of these alleles.
HRM analysis of hsp70 real-time PCR products corroborated the RFLP results
The evaluation of polymorphic nucleotide sequences by HRM analysis revealed differences between the parental and hybrid haplotypes in five putative hybrids isolates (1, 2, 3, 4 and 5). One of the main parameters used in the analysis was the melting temperature (Tm). Discrete variations in the nucleotide composition of DNA fragments are reflected in Tm variations. The Tm values for the L. (V.) shawi and L. (V.) guyanensis hsp70 PCR products were 86.80 °C and 86.14 °C, respectively (Figure 2A and Figure 2B). The hybrid parasites with a homozygous hsp70 profile showed a dissociation temperature and melting curve corresponding to L. (V.) shawi (Figure 2A and Figure 2B). The hsp70 heterozygous hybrids showed a unique dissociation temperature and a melting curve distinct from those of all other examined species.
HRM using hsp70 as target. A- Representative dispersion graph shows the average and standard deviation of the Tm values. B-Normalized melting curves for L. (V.) guyanensis, L. (V.) shawi and hybrid isolates 1, 2, 3, 4 and 5.
Discussion
The strains characterized in the present study were isolated from patients inhabiting the lower Amazon region in Pará State, an important endemic area of leishmaniasis in Brazil. The identification of seven different Leishmania strains involved in ACL presenting high intraspecific diversity in this geographic area is relevant to epidemiological studies (Jennings et al., 2014Jennings YL, Souza AAA, Ishikawa EA, Shaw JJ, Lainson R and Silveira FT (2014) Phenotypic characterization of Leishmania spp causing cutaneous leishmaniasis in the lower Amazon region, Western Pará State, Brazil, reveals a putative hybrid parasite, Leishmania (Viannia) guyanensis×Leishmania (Viannia) shawi shawi. Parasite 21:39.; de Souza et al., 2016de Souza ACA, dos Santos TV, Jennings YLL, Ishikawa EAY, Barata IR, Silva MGS, Lima JAN, Shaw J, Lainson R and Silveira FT (2016) Natural Leishmania (Viannia) spp. infections in Phlebotomine sand flies (Diptera: Psychodidae) from the Brazilian Amazon region reveal new putative transmission cycles of american cutaneous leishmaniasis. Parasite 23:22.).
Santarém is also an important endemic area of visceral leishmaniasis (Braga et al., 1986Braga RR, Lainson R, Shaw JJ, Ryan L and Silveira FT (1986) Leishmaniasis in Brazil. XXII: characterization of Leishmania from man, dogs and the sandfly Lutzomyia longipalpis (Lutz & Neiva, 1912) isolated during an outbreak of visceral leishmaniasis in Santarém, Pará state. Trans R Soc Trop Med Hyg 80:143-145.). The considerable diversity of vectors and reservoirs in the Amazon rain forest, one of the most complex, diverse biomes on the planet, is also notable (Christensen et al., 1982Christensen HA, Arias JR, De Vasquez AM and de Freitas RA (1982) Hosts of sandfly vectors of Leishmania braziliensis guyanensis in the Central Amazon of Brazil. Am J Trop Med Hyg 31:239-242.; Grimaldi et al., 1991Grimaldi GJ, Momen H, Naiff RD, McMahon-Pratt D and Barrett TV (1991) Characterization and classification of leishmanial parasites from humans, wild mammals, and sand flies in the amazon region of Brazil. Am J Trop Med Hyg 44:645-661.; Silveira et al., 1991Silveira FT, Souza AA, Lainson R, Shaw JJ, Braga RR and Ishikawa EE (1991) Cutaneous leishmaniasis in the Amazon region:natural infection of sandfly Lutzomyia ubiquitalis (Psychodidae: Phlebotominae) by Leishmnaia (Viannia) lainsoni in Pará state, Brazil. Mem Inst Oswaldo Cruz 86:127-130.; Lainson et al., 1994Lainson R, Shaw JJ, Silveira FT, de Souza AA, Braga RR and Ishikawa EA (1994) The dermal leishmaniases of Brazil, with special reference to the eco-epidemiology of the disease in Amazonia. Mem Inst Oswaldo Cruz 89:435-443.).
Due to the species diversity observed in Santarém, each isolate was initially cloned to eliminate the possibility of a mixed infection.
The patterns of 234 bp fragments from hsp70 PCR-RFLP analysis yielded two profiles. One was similar to that of the parental species L. (V.) shawi, which was considered a homozygous pattern. The other pattern showed bands characteristic of both L. (V.) shawi and L. (V.) guyanensis and was considered a heterozygous profile, which is commonly used parameter to confirm hybridization events between species (Gaunt et al., 2003Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, Taylor MC, Mena SS, Veazey P, Miles GAJ, Acosta N, et al. (2003) Mechanism of genetic exchange in american trypanosomes. Nature 421:936-939.; Inbar et al., 2013Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, Kauffmann F, Barhoumi M, Grigg M, Owens K, et al. (2013) The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672.). According to studies that have investigated Leishmania mating, the inheritance profile of the parasite consists of the same number of chromosomes derived from each parent during a sexual reproduction event, generating heterozygous hybrids (Akopyants et al., 2009Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM and Sacks DL (2009) Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324:265-268.; Rogers et al., 2014Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA and Smith DF (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092.). The PCR-RFLP analysis performed for mpi alleles showed heterozygous or homozygous profiles for L. (V.) shawi or L. (V.) guyanensis. These findings reinforced the previous phenotypic findings and, together with the hsp70 results, confirmed the hybrid nature of these isolates.
The isolates 4 and 7 presented hsp70 and mpi alleles compatible with L. (V.) shawi (Table S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. ). However, isoenzyme analyses showed that these strains harbor at least two isoenzymes compatible with L. (V.) guyanensis, which strongly suggest the hypothesis of their hybrid nature, as demonstrated in Table S1. In addition, the three band profile observed for the dimeric 6PGD isoenzyme via MLEE analysis was associated with a heterozygous hybrid profile (Jennings et al., 2014Jennings YL, Souza AAA, Ishikawa EA, Shaw JJ, Lainson R and Silveira FT (2014) Phenotypic characterization of Leishmania spp causing cutaneous leishmaniasis in the lower Amazon region, Western Pará State, Brazil, reveals a putative hybrid parasite, Leishmania (Viannia) guyanensis×Leishmania (Viannia) shawi shawi. Parasite 21:39.). Similar findings were reported for L. (V.) braziliensis /L. (V.) guyanensis hybrids from Venezuela (Dujardin et al., 1995Dujardin JC, Bañuls AL , Llanos-Cuentas A, Alvarez E, DeDoncker S, Jacquet D, Le Ray D, Arevalo J and Tibayrenc M (1995) Putative Leishmania hybrids in the eastern Andean valley of Huanuco, Peru. Acta Trop 59:293-307.; Delgado et al., 1997Delgado O, Cupolillo E, Bonfante-Garrido R, Silva S, Belfort E, Grimaldi G Júnior and Momen H (1997) Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem Inst Oswaldo Cruz 92:581-582.). Rogers et al. (2014Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA and Smith DF (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092.) proposed that after an initial mating event, the hybrid cells participate in inbreeding events exclusively among themselves. This hypothesis could justify the homozygous profile found for some isolates because a high endogamy rate can lead to loss of heterozygosity (Inbar et al., 2013Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, Kauffmann F, Barhoumi M, Grigg M, Owens K, et al. (2013) The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672.; Gelanew et al., 2014Gelanew T, Hailu A, Schőnian G, Lewis MD, Miles MA and Yeo M (2014) Multilocus sequence and microsatellite identification of intra-specific hybrids and ancestor-like donors among natural ethiopian isolates of Leishmania donovani. Int. J. Parasitol 44:751-757.; Rogers et al., 2014Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA and Smith DF (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092.). Another important aspect that must be considered in relation to the homozygous profile is mosaic aneuploidy, a widespread characteristic of the genus Leishmania (Rogers et al., 2011Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledje DP, Harris D, Her Y, Herzyk P, Imamura H, et al. (2011) Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21:2129-2142.; Sterkers et al., 2012Sterkers Y, Lachaud L, Bourgeois N, Crobu L, Bastien P and Pagès M (2012) Novel insights into genome plasticity in eukaryotes: mosaic aneuploidy in Leishmania. Mol Microbiol 86:15-23.). This phenomenon is probably caused by a chromosomal replication defect, followed by asymmetric segregation that occurs rapidly and that leads to the loss of heterozygosity (Sterkers et al., 2011Sterkers Y, Lachaud L, Crobu L, Bastien P and Pagès M (2011) FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol 13:274-283.; Inbar et al., 2013Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, Kauffmann F, Barhoumi M, Grigg M, Owens K, et al. (2013) The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672.). This factor cannot be ignored because hybrid strains are established by clonal propagation and are therefore subject to the events associated with such mechanisms.
Four isolates presented a heterozygous hybrid profile for at least one analyzed molecular target and one isolate presented hsp70 heritage from L. (V.) shawi and mpi from L. (V.) guyanensis. Altogether, these molecular analyses confirmed that at least five isolates are true L. (V.) shawi and L. (V.) guyanensis hybrids.
Considering the variable isoenzyme plus hsp70 and mpi RFLP profiles, the seven isolates were divided into six different groups (Table S1 Table S1 - Compiled data of hsp70 and mpi PCR RFLP and multilocus enzyme electrophoresis (MLEE) from Jennings et al., 2014. ). Such diversity among hybrid isolates is supported by the hybridization and recombination model proposed by Rogers et al. (2014Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA and Smith DF (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092.). This could suggest that hybridization events may be advantageous for a genus, offsetting the predominant mode of clonal reproduction to some extent. It is also notable interesting to note that different hybrid strains were found in patients with ACL, reflecting a well-established cycle in the environment. More information about the niche occupied by different hybrids in the environment would help to elucidate the interactions among the various genotypes and their establishment in the environment. A previous study suggested that Nyssomyia whitmani can act as permissive vector, in which mating events between L. (V.) shawi and L. (V.) guyanensis could occur (de Souza et al., 2016de Souza ACA, dos Santos TV, Jennings YLL, Ishikawa EAY, Barata IR, Silva MGS, Lima JAN, Shaw J, Lainson R and Silveira FT (2016) Natural Leishmania (Viannia) spp. infections in Phlebotomine sand flies (Diptera: Psychodidae) from the Brazilian Amazon region reveal new putative transmission cycles of american cutaneous leishmaniasis. Parasite 23:22.).
The SNP-CAPS analysis showed a similar intensity between the putative homozygous hsp70 hybrids and the L. (V.) shawi hsp70 fragment, suggesting that they have the same number of copies of this gene. In relation to the heterozygous hsp70 profile, the SNP-CAPS assay suggested that L. (V.) guyanensis hsp70 copies are more heavily represented in the genotypes of the hybrids. The comparative analysis of the relative intensity between the 180 bp band from L. (V.) guyanensis and the 120 bp band from L. (V.) shawi showed that the heterozygous isolates presented a L. (V.) guyanensis band that was approximately 6x more intense. The 5 and 6 isolates presented three homozygous isoenzyme profiles, two of which were similar to that of L. (V.) guyanensis. The 3 isolate presented all the isoenzymes of a homozygous profile similar to that of L. (V.) guyanensis. These findings suggest that L. (V.) guyanensis accounts for a higher proportion of the phenotypic expression of the hybrids. However, other tests are necessary to confirm this hypothesis.
It is important to emphasize that different L. (V.) shawi and L. (V.) guyanensis strains from Pará State and the North region of Brazil included in our analysis (Table 1) presented the same RFLP and HRM profile, demonstrating that these targets are conserved among the strains used in this study, even between L. (V.) shawi shawi and L. (V.) shawi santarensis subpopulations. However, previous findings have suggested that the L. (V.) shawi shawi subpopulation is the probable the parental species (de Souza et al., 2016de Souza ACA, dos Santos TV, Jennings YLL, Ishikawa EAY, Barata IR, Silva MGS, Lima JAN, Shaw J, Lainson R and Silveira FT (2016) Natural Leishmania (Viannia) spp. infections in Phlebotomine sand flies (Diptera: Psychodidae) from the Brazilian Amazon region reveal new putative transmission cycles of american cutaneous leishmaniasis. Parasite 23:22.).
The present study confirmed by molecular methods, the existence of at least five strains of hybrid parasites L. (V.) shawi / L. (V.) guyanensis involved in cases of ATL in the Amazon region of Santarém Para, Brazil.
Acknowledgments
The authors would like to thank the Laboratorio de Patologia de Molestias Infecciosas (LIM50 HCFMUSP), Departamento de Patologia, Faculdade de Medicina FMUSP, Universidade de Sao Paulo, Sao Paulo, SP, BR; Laboratório de Leishmanioses "Prof. Dr. Ralph Lainson" do Instituto Evandro Chagas; Laboratório de Fisiologia de Trypanosomatideos (IB-USP); Dr. André André G. Costa-Martins from Universidade de Sao Paulo for helping to submit the sequences in NCBI and Dr. Emerson Castilho from the Universidade Federal do Amapá for review of the manuscript. The work was supported by FAPESP (2012/08338-8; 14/50315-0).
References
- Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM and Sacks DL (2009) Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324:265-268.
- Bastien P, Blaineau C and Pages M (1992) Leishmania: sex, lies and karyotype. Parasitol Today 8:174-177.
- Boité MC, Mauricio IL, Miles MA and Cupolillo E (2012) New insights on taxonomy, phylogeny and population genetics of Leishmania (Viannia) parasites based on multilocus sequence analysis. Plos Negl Trop Dis 6:e1888.
- Braga RR, Lainson R, Shaw JJ, Ryan L and Silveira FT (1986) Leishmaniasis in Brazil. XXII: characterization of Leishmania from man, dogs and the sandfly Lutzomyia longipalpis (Lutz & Neiva, 1912) isolated during an outbreak of visceral leishmaniasis in Santarém, Pará state. Trans R Soc Trop Med Hyg 80:143-145.
- Christensen HA, Arias JR, De Vasquez AM and de Freitas RA (1982) Hosts of sandfly vectors of Leishmania braziliensis guyanensis in the Central Amazon of Brazil. Am J Trop Med Hyg 31:239-242.
- Delgado O, Cupolillo E, Bonfante-Garrido R, Silva S, Belfort E, Grimaldi G Júnior and Momen H (1997) Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis Mem Inst Oswaldo Cruz 92:581-582.
- Dujardin JC, Bañuls AL , Llanos-Cuentas A, Alvarez E, DeDoncker S, Jacquet D, Le Ray D, Arevalo J and Tibayrenc M (1995) Putative Leishmania hybrids in the eastern Andean valley of Huanuco, Peru. Acta Trop 59:293-307.
- Dujardin JC, De Doncker S, Jacquet D, Bañuls AL, Balavoine M, van Bockstaele D, Tibayrenc M, Arevalo J and Le Ray D (2007) Clonal propagation and the fast generation of karyotype diversity: an in vitro Leishmania model. Parasitology 134:33-39.
- Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, Taylor MC, Mena SS, Veazey P, Miles GAJ, Acosta N, et al (2003) Mechanism of genetic exchange in american trypanosomes. Nature 421:936-939.
- Gelanew T, Hailu A, Schőnian G, Lewis MD, Miles MA and Yeo M (2014) Multilocus sequence and microsatellite identification of intra-specific hybrids and ancestor-like donors among natural ethiopian isolates of Leishmania donovani Int. J. Parasitol 44:751-757.
- Graça GC, Volpini AC, Romero GAS, Oliveira Neto MP, Hueb M, Porrozzi R, Boité MC and Cupolillo E (2012) Development and validation of PCR-based assays for diagnosis of american cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz 107:664-674.
- Grimaldi GJ, Momen H, Naiff RD, McMahon-Pratt D and Barrett TV (1991) Characterization and classification of leishmanial parasites from humans, wild mammals, and sand flies in the amazon region of Brazil. Am J Trop Med Hyg 44:645-661.
- Handman E, Hocking RE, Mitchell GF and Spithill TW (1983) Isolation an characterization of infective and non-infective clones of Leishmania tropica Mol Biochem Parasitol 7:111-126.
- Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, Kauffmann F, Barhoumi M, Grigg M, Owens K, et al (2013) The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672.
- Inbar E, Shaik J, Iantorno SA, Romano A, Nzelu CO, Sanders MJ, Dobson D, Cotton JA, Grigg ME, Beverley SM, et al (2019) Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania PLoS Genet 15:e1008042.
- Jennings YL, Souza AAA, Ishikawa EA, Shaw JJ, Lainson R and Silveira FT (2014) Phenotypic characterization of Leishmania spp causing cutaneous leishmaniasis in the lower Amazon region, Western Pará State, Brazil, reveals a putative hybrid parasite, Leishmania (Viannia) guyanensis×Leishmania (Viannia) shawi shawi Parasite 21:39.
- Kato H, Gomez EA, Seki C, Furumoto H, Martini-Hobles L, Muzzio J, Calvopiña M, Velez L, Kubo M, Tabbabi A, et al (2019) PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. Plos Negl Trop Dis 13:e0007403.
- Lainson R, Shaw JJ, Silveira FT, de Souza AA, Braga RR and Ishikawa EA (1994) The dermal leishmaniases of Brazil, with special reference to the eco-epidemiology of the disease in Amazonia. Mem Inst Oswaldo Cruz 89:435-443.
- Muniaraj M, Sinha PK and Das P (2010) Antileishmanial activity of drug infused mini-agar plates on Leishmania donovani promastigotes. Trop Biomed 27:657-661.
- Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledje DP, Harris D, Her Y, Herzyk P, Imamura H, et al (2011) Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania Genome Res 21:2129-2142.
- Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA and Smith DF (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092.
- Rougeron V, Meeûs TD, Hide M, Waleckx E, Bermudez H, Arevalo J, Llanos-Cuentas A, Dujardin JC, De Doncker S, Le Ray D, et al (2009) Extreme inbreeding in Leishmania braziliensis PNAS 106:10224-10229.
- Silveira FT, Souza AA, Lainson R, Shaw JJ, Braga RR and Ishikawa EE (1991) Cutaneous leishmaniasis in the Amazon region:natural infection of sandfly Lutzomyia ubiquitalis (Psychodidae: Phlebotominae) by Leishmnaia (Viannia) lainsoni in Pará state, Brazil. Mem Inst Oswaldo Cruz 86:127-130.
- de Souza ACA, dos Santos TV, Jennings YLL, Ishikawa EAY, Barata IR, Silva MGS, Lima JAN, Shaw J, Lainson R and Silveira FT (2016) Natural Leishmania (Viannia) spp. infections in Phlebotomine sand flies (Diptera: Psychodidae) from the Brazilian Amazon region reveal new putative transmission cycles of american cutaneous leishmaniasis. Parasite 23:22.
- Sterkers Y, Lachaud L, Crobu L, Bastien P and Pagès M (2011) FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major Cell Microbiol 13:274-283.
- Sterkers Y, Lachaud L, Bourgeois N, Crobu L, Bastien P and Pagès M (2012) Novel insights into genome plasticity in eukaryotes: mosaic aneuploidy in Leishmania Mol Microbiol 86:15-23.
- Tibayrenc M and Ayala FJ (1991) Towards a population genetics of microorganisms: the clonal theory of parasitic protozoa. Parasitol Today 7:228-232.
- Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Brenière SF, Dardé ML and Ayala FJ (1991) Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci U S A 88:5129-5133.
- Uliana SR, Affonso MH, Camargo EP and Floeter-Winter LM (1991) Leishmania: genus identification based on a specific sequence of the 18S ribosomal RNA sequence. Exp Parasitol 72:157-163.
- Victoir K and Dujardin JC (2002) How to succeed in parasitic life without sex? Asking Leishmania Trends Parasitol 18:81-85.
- Zampieri RA, Laranjeira-Silva MF, Muxel SM, Lima ACS, Shaw JJ and Floeter-Winter LM (2016) High Resolution Melting Analysis targeting hsp70 as a fast and efficient method for the discrimination of Leishmania species. PLoS Negl Trop Dis 10:e0004485.
Supplementary material
The following online material is available for this article:
Table S2 - NCBI sequences accession numbers.
Figure S2 - Alignment of 234 bp fragments used in hsp70-RFLP approach.
Publication Dates
-
Publication in this collection
30 Apr 2021 -
Date of issue
2021
History
-
Received
23 Apr 2020 -
Accepted
03 Mar 2021