Abstract
In this study, 100 clinical isolates of Streptococcus agalactiae recovered from genitourinary tract specimens of non-pregnant individuals living in Rio de Janeiro were submitted for antimicrobial susceptibility testing, detection of macrolide resistance genes and evaluation of the genetic diversity of erythromycin-resistant isolates. By agar diffusion method, all isolates were susceptible to ceftazidime, penicillin and vancomycin. Isolates were resistant to levofloxacin (1%), clindamycin (5%), erythromycin (11%) and tetracycline (83%) and were intermediated to erythromycin (4%) and tetracycline (6%). Erythromycin-resistant and intermediated isolates presented the following phenotypes: M (n = 3), constitutive macrolide-lincosamide-streptogramin B (MLS B, n = 5) and inductive MLS B (n = 7). Determinants of macrolide resistance genes, erm and mef, were detected in isolates presenting MLS B and M phenotypes, respectively. Randomly amplified polymorphic DNA profiles of erythromycin-resistant isolates were clustered into two major groups of similarity.
Streptococcus agalactiae; antimicrobial resistance; erythromycin resistance genes
ARTICLES
Antimicrobial resistance profiles and genetic characterisation of macrolide resistant isolates of Streptococcus agalactiae
Priscila AM NakamuraI; Rôde Beatriz B SchuabI; Felipe PG NevesI; Cláudio FA PereiraI; Geraldo R de PaulaII; Rosana R BarrosI, + + Corresponding author: miprosana@vm.uff.br
IDepartamento de Microbiologia e Parasitologia, Instituto Biomédico
IIDepartamento de Tecnologia Farmacêutica, Faculdade de Farmácia, Universidade Federal Fluminense, Rua Prof. Hernani de Melo 101, 24210-130 Niterói, RJ, Brasil
ABSTRACT
In this study, 100 clinical isolates of Streptococcus agalactiae recovered from genitourinary tract specimens of non-pregnant individuals living in Rio de Janeiro were submitted for antimicrobial susceptibility testing, detection of macrolide resistance genes and evaluation of the genetic diversity of erythromycin-resistant isolates. By agar diffusion method, all isolates were susceptible to ceftazidime, penicillin and vancomycin. Isolates were resistant to levofloxacin (1%), clindamycin (5%), erythromycin (11%) and tetracycline (83%) and were intermediated to erythromycin (4%) and tetracycline (6%). Erythromycin-resistant and intermediated isolates presented the following phenotypes: M (n = 3), constitutive macrolide-lincosamide-streptogramin B (MLSB, n = 5) and inductive MLSB (n = 7). Determinants of macrolide resistance genes, erm and mef, were detected in isolates presenting MLSB and M phenotypes, respectively. Randomly amplified polymorphic DNA profiles of erythromycin-resistant isolates were clustered into two major groups of similarity.
Key words: Streptococcus agalactiae - antimicrobial resistance - erythromycin resistance genes
Streptococcus agalactiae [β-haemolytic group B streptococci (GBS)] is a component of human intestinal and genitourinary microflora. This species is frequently related to illness in newborns, causing life-threatening diseases such as meningitis and bacteremia and is associated with complications during pregnancy and the post-partum period. GBS colonisation of the maternal genital tract has been considered the most important risk factor for neonatal disease development (Schuchat & Wenger 1994). GBS is also responsible for infections in older infants and non-pregnant adults, especially the elderly or those with any underlying medical conditions. Clinical manifestations include urinary tract infections, skin or soft tissue infections, osteomyelitis and bacteremia (Sendi et al. 2008). Points of concern include the increasing frequency of infections outside the perinatal period and a higher proportion of mortality in adults compared to the neonatal group (Phares et al. 2008).
GBS has been continuously susceptible to penicillin and other β-lactams. However, resistance to antimicrobials used as alternative therapy, especially macrolides, lincosamides and fluoroquinolones has been documented in different countries (Duarte et al. 2005, Tazi et al. 2008, Pinheiro et al. 2009). One of the most common macrolide resistance mechanisms in streptococci is ribosomal modification by methylases, which are encoded by the erm genes. These enzymes also confer inducible or constitutive resistance to lincosamides and streptogramin B, which characterises macrolide-lincosamide-streptogramin B (MLSB) phenotypes. Another common mechanism is drug efflux by a membrane-bound protein encoded by the mef gene, which is associated with the M phenotype (de Azavedo et al. 2001). Resistance to fluoroquinolones in GBS was first described in 2003 and is associated with point mutations in gyrA and parC genes (Kawamura et al. 2003).
Despite the clinical impact of GBS infections and increasing resistance rates to some antimicrobials, there are a limited number of studies reporting antimicrobial resistance profiles among GBS strains circulating in Brazil, even from asymptomatic colonisation or clinical infection. In this study, 100 strains of S. agalactiae, isolated from genitourinary tract specimens from non-pregnant individuals during January 2008-August 2009 in the metropolitan area of Rio de Janeiro, were tested for antimicrobial susceptibility and the presence of macrolide resistance genes. The genetic diversity of erythromycin resistant isolates was evaluated using randomly amplified polymorphic-polymerase chain reaction (RAPD-PCR).
SUBJECTS, MATERIALS AND METHODS
Strains - One hundred S. agalactiae isolates from genitourinary tract specimens processed in a clinical laboratory in the metropolitan area of Rio de Janeiro during January 2008-August 2009 were included this study. The specimens [vaginal secretions (n = 49), urine (n = 30), sperm (n = 17) and other secretions (n = 4)] were obtained from non-pregnant individuals (73 females and 27 males, with ages ranging from 6-79 years). Conventional tests (detection of CAMP factor, hippurate hydrolysis and streptococcal serogrouping) were used to confirm species identification, as previously described (Ruoff et al. 1999).
Antimicrobial susceptibility testing - All isolates were submitted to susceptibility tests to ceftriaxone (30 μg), clindamycin (2 μg), erythromycin (15 μg), levofloxacin (5 μg), penicillin (10 U), tetracycline (30 μg) and vancomycin (30 μg) (Cecon, São Paulo) by agar diffusion method, according to CLSI (2009b, c). Azithromycin (15 μg) and ciprofloxacin (5 μg) disks were tested on erythromycin and levofloxacin-resistant isolates, respectively. Macrolide resistance phenotypes were detected by double-disk test with erythromycin (15 μg) and clindamycin (2 μg) disks. Erythromycin and levofloxacin minimal inhibitory concentration (MIC) determination was performed on resistant isolates using agar dilution (CLSI 2009a, c) and E-test methods, respectively.
Detection of erythromycin resistance genes - PCR was used to detect ermA, ermB and mefA/E genes in erythromycin-resistant isolates. The DNA template was prepared as described by Beall et al. (1996), except for the fact that hyaluronidase solution was not used. PCR reagents, agarose and DNA marker were obtained from Fermentas (Burlington, ON, Canada). Primers were described elsewhere (Pérez-Trallero et al. 2007). The reaction mixtures, in final volumes of 30 μL, contained MgCl2 (2 mM), dNTPs (0.2 mM each), primers (1 μM each), Taq DNA polymerase (1 U), reaction buffer (10 mM), and 3 μL of DNA template. Cycling was performed in the GeneAmp 9700 Thermocycler (Applied Biosystems, Foster City, CA, USA), as follows: initial denaturation at 94ºC for 3 min, followed by 35 cycles of denaturation at 94ºC for 1 min, annealing at 50ºC for 1 min and elongation at 72ºC for 1 min. A final extension step was carried out at 72ºC for 5 min. PCR products were resolved on 1.2% agarose gels.
RAPD-PCR - DNA polymorphisms of all macrolide resistant and 10 susceptible isolates were obtained by RAPD-PCR using primer 1254 (5'-CCGCAGCCAA-3'). Reaction mixtures and PCR conditions were the same used to investigate erythromycin resistance genes, except the temperature of the annealing step was changed to 37ºC. PCR products were resolved on 1.2% agarose gels. RAPD profiles were analysed using the Molecular Analyst Fingerprint Plus software package v1.12 (Bio-Rad, Hercules, CA, USA).
RESULTS
All isolates hydrolysed sodium hippurate and CAMP factor was detected in 98% of isolates. CAMP-negative isolates agglutinated with streptococcal group B antiserum. By disk diffusion method, all isolates were susceptible to ceftriaxone, penicillin and vancomycin. Isolates were resistant to tetracycline (83%), erythromycin (11%), clindamycin (5%) and levofloxacin (1%) and intermediate to tetracycline (6%) and erythromycin (4%). Levofloxacin resistance in a unique isolate was confirmed by MIC determination (> 32 μg/mL). This isolate was also resistant to ciprofloxacin by disk diffusion.
Erythromycin resistant and intermediate isolates were also resistant to azithromycin and tetracycline and presented the following phenotypes: inducible MLSB (iMLSB) (n = 7), constitutive MLSB (cMLSB) (n = 5) and M (n = 3). Erythromycin MIC values varied from 0.5 to >256 μg/mL. Higher MIC values were observed in cMLSB isolates. Three isolates, intermediate by disk diffusion, were considered resistant by agar dilution method. Considering that the MIC determination is the standard method for susceptibility testing, the overall rate of erythromycin resistance was 14%.
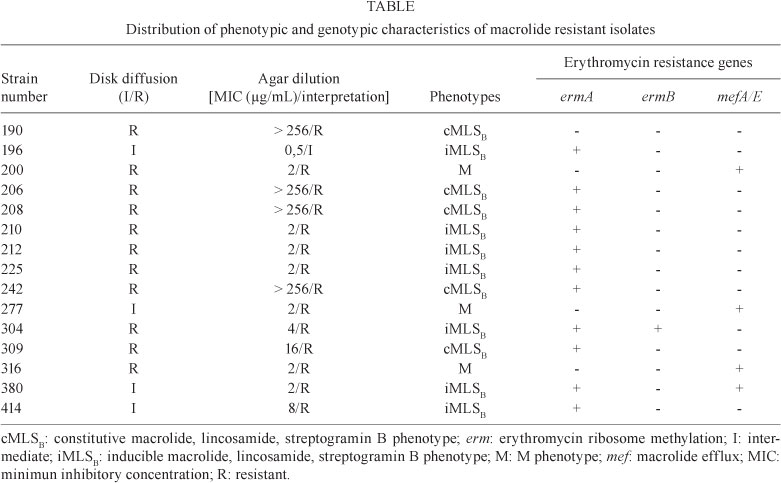
Investigation of macrolide resistance genes generated PCR products typical of ermA (375 bp), ermB (442 bp) or mefA/E (345 bp) in 14 isolates. The mefA/E gene was detected in all M phenotype isolates and in one isolate that presented an iMLSB phenotype. The ermA gene was detected in all isolates that presented iMLSB and cMLSB phenotypes, while ermB was detected in only one iMLSB isolate. Two iMLSB isolates harboured a combination of genes (ermA + ermB and ermA + mefA/E). None of the investigated genes were detected in one isolate exhibiting cMLSB phenotype. Despite the failure to amplify erm or mef genes, an amplicon compatible to the 16S DNA gene was obtained in a PCR performed to check the quality of reagents and DNA template of this isolate (data not shown). Phenotypic and genotypic characteristics of macrolide resistant isolates are shown in Table.
All erythromycin-resistant or intermediate and 10 representative erythromycin-susceptible isolates, including a unique levofloxacin-resistant isolate, were submitted to evaluation of genetic relatedness by RAPD-PCR. A variety of reproducible profiles, comprising up to eight bands, were generated. By computer-assisted analysis, isolates were clustered in two major groups, with different levels of similarity in each one. The relationships among RAPD profiles are represented in Figure.
DISCUSSION
Resistance to erythromycin, clindamycin and tetracycline was observed among the studied isolates. GBS resistance rates to such antimicrobials have varied according to the area and period evaluated, as shown in studies conducted in different countries (de Azavedo et al. 2001, Alhambra et al. 2004, Pinheiro et al. 2009, Ulett et al. 2009). In Brazil, most of the available data about GBS antimicrobial susceptibility was obtained from studies performed in Rio de Janeiro (d'Oliveira et al. 2003, Borger et al. 2005, Duarte et al. 2005). In this study, tetracycline and clindamycin resistance rates were very similar to those observed in previous studies conducted in the same area.
A point of concern is the finding of a levofloxacin-resistant isolate. GBS highly resistant to fluoroquinolones was first described in Japan in 2003 (Kawamura et al. 2003) and, since then, it has been reported in other countries (Miró et al. 2006, Tazi et al. 2008). To our knowledge, this report is the first of fluoroquinolone resistance among GBS circulating in Brazil.
Regarding resistance to erythromycin, the alternative therapy to penicillin allergic patients, results obtained in this study far exceeded the rates previously observed in the same geographical area. Considering the period in which each study was performed, it is possible to conclude that resistance to erythromycin in the metropolitan area of Rio de Janeiro has risen from 5.4% (d'Oliveira et al. 2003) to 9.4% (Borger et al. 2005) to 14%, as observed in this study. Although there is no available data on macrolide consumption in this area, it is well documented that, for related species, resistance to this antimicrobial is directly linked to its use (Seppälä et al. 1997). Agar dilution and disk diffusion methods yielded concordant results to all isolates except for three that were intermediate by disk diffusion and resistant by agar dilution. Such discrepancies when testing erythromycin were previously reported in GBS isolates (d'Oliveira et al. 2003). Macrolide resistance phenotypes were well correlated with genotypes, except for one isolate that exhibited a cMLSB phenotype and did not harbour any of the investigated genes. The failure to detect erm and mef genes in cMLSB phenotype isolates has been described elsewhere (Pinheiro et al. 2009). Combination of genetic determinants ermA + ermB in isolates that expressed an iMLSB phenotype has been described in different areas, including Brazil (Duarte et al. 2005, Pinheiro et al. 2009). However, combination of ermA + mefA/E genes in an iMLSB phenotype had never been described in erythromycin-resistant isolates circulating in Brazil.
RAPD-PCR is a technique that evaluates overall DNA polymorphisms using random oligonucleotides and has been applied to evaluate genetic diversity of GBS (Duarte et al. 2004, Hannoun et al. 2009). RAPD profiles obtained from erythromycin-resistant isolates generated a dendrogram with two major clusters, with different levels of similarity in each one. A few clusters that comprised isolates with significant genetic variability were also observed in a study by Duarte et al. (2004), which applied RAPD-PCR and pulsed field gel electrophoresis (PFGE) to type bovine GBS. The observed variability may reflect multiclonal spread of genetic determinants of resistance, instead of dissemination of a few clones. According to the authors, although PFGE has been considered the gold standard technique to type many bacterial species, isolates were clustered in a very similar way by both methodologies, reflecting the reliability of RAPD-PCR as an alternative typing system for GBS. Additionally, resistant and susceptible erythromycin isolates were clustered together in each major group. Similar results were reported in a recent study that used RAPD-PCR to evaluate genetic relatedness of GBS submitted for antimicrobial susceptibility testing (Hannoun et al. 2009). These results suggest that RAPD methodology, despite its limitations as an approach to evaluate genetic relatedness, was sufficiently reliable to exclude the presence of a single clonal group among erythromycin resistant isolates.
In conclusion, this study highlights the erythromycin resistance rate, novel combinations of macrolide resistance genetic determinants and the occurrence of levofloxacin resistance among S. agalactiae circulating in Rio de Janeiro. Knowledge of current antimicrobial susceptibility profiles and genetic characteristics of GBS in different geographic areas is essential to the development of strategies that focus on the prevention and treatment of streptococcal infections.
ACKNOWLEDGEMENTS
To Laboratórios Daflon, Helion Póvoa and Maiolino, for strains donation.
Received 11 June 2010
Accepted 25 January 2011
Financial support: FAPERJ (APQ1 170.261/2006), PROPPi-UFF
- Alhambra A, Gómez-Garcés JL, Alós JI 2004. Susceptibility of Streptococcus agalactiae isolates from blood and urine to 18 widely used and recently marketed antibiotics. Clin Microbiol Infect 10: 267-268.
- Beall B, Facklam R, Thompson T 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol 34: 953-958.
- Borger IL, D'Oliveira REC, Castro ACD, Mondino SSB 2005. Streptococcus agalactiae em gestantes: prevalência de colonização e avaliação de suscetibilidade aos antimicrobianos. Rev Bras Ginecol Obstet 27: 575-579.
- CLSI - Clinical Laboratory Standard Institute 2009a. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard - eighth edition, M07-A8, Clinical Laboratory Standard Institute, Wayne, 10 pp.
- CLSI - Clinical Laboratory Standard Institute 2009b. Performance standards for antimicrobial disk susceptibility tests; approved standard - tenth edition, M02-A10, Clinical Laboratory Standard Institute, Wayne, 10 pp.
- CLSI - Clinical Laboratory Standard Institute 2009c. Performance standards for antimicrobial susceptibility testing, 19th informational supplement, M100-S19, Clinical Laboratory Standard Institute, Wayne, 10 pp.
- de Azavedo JC, McGavin M, Duncan C, Low DE, McGeer A 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob Agents Chemother 45: 3504-3508.
- d'Oliveira RE, Barros RR, Mendonça CR, Teixeira LM, Castro AC 2003. Susceptibility to antimicrobials and mechanisms of erythromycin resistance in clinical isolates of Streptococcus agalactiae from Rio de Janeiro, Brazil. J Med Microbiol 52: 1029-1030.
- Duarte RS, Bellei BC, Miranda OP, Brito MA, Teixeira LM 2005. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob Agents Chemother 49: 97-103.
- Duarte RS, Miranda OP, Bellei BC, Brito MA, Teixeira LM 2004. Phenotypic and molecular characteristics of Streptococcus agalactiae isolates recovered from milk of dairy cows in Brazil. J Clin Microbiol 42: 4214-4222.
- Hannoun A, Shehab M, Khairallah MT, Sabra A, Abi-Rached R, Bazi T, Yunis KA, Araj GF, Matar GM 2009. Correlation between group B streptococcal genotypes, their antimicrobial resistance profiles and virulence genes among pregnant women in Lebanon. Int J Microbiol 2009: 1-6.
- Kawamura Y, Fujiwara H, Mishima N, Tanaka Y, Tanimoto A, Ikawa S, Itoh Y, Ezaki T 2003. First Streptococcus agalactiae isolates highly resistant to quinolones with point mutations in gyrA and parC. Antimicrob Agents Chemother 47: 3605-3609.
- Miró E, Rebollo M, Rivera A, Álvarez MT, Navarro F, Mirelis B, Coll P 2006. Streptococcus agalactiae altamente resistente a fluoroquinolonas. Enferm Infec Microbiol Clin 24: 562-563.
- Pérez-Trallero E, Montes M, Orden B, Tamayo E, García-Arenzana JM, Marimón JM 2007. Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob Agents Chemother 51: 1228-1233.
- Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ, Active Bacterial Core surveillance/Emerging Infections Program Network 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299: 2056-2065.
- Pinheiro S, Radhouani H, Coelho C, Gonçalves A, Carvalho E, Carvalho JA, Ruiz-Larrea F, Torres C, Igrejas G, Poeta P 2009. Prevalence and mechanisms of erythromycin resistance in Streptococcus agalactiae from healthy pregnant women. Microb Drug Resist 15: 121-124.
- Ruoff KL, Whiley RA, Beighton D 1999. Streptococcus In PR Murray, EJ Baron, MA Pfaller, FC Tenover, RH Yolken, Manual of clinical microbiology, 7th ed., American Society for Microbiology, Washington DC, p. 283-296.
- Schuchat A, Wenger JD 1994. Epidemiology of group B streptococcal disease. Risk factors, prevention strategies and vaccine development. Epidemiol Rev 16: 374-402.
- Sendi P, Johansson L, Norrby-Teglund A 2008. Invasive group B streptococcal disease in non-pregant adults: a review with emphasis on skin and soft-tissue infections. Infection 36: 100-111.
- Seppälä H, Klaukka T, Vuopio-Varkila J, Muotiala A, Helenius H, Lager K, Huovinen P, Finnish Group for Antimicrobial Resistance 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N Engl J Med 337: 441-446.
- Tazi A, Gueudet T, Varon E, Gilly L, Trieu-Cuot P, Poyart C 2008. Fluoroquinolone-resistant group B streptococci in acute exacerbation of chronic bronchitis. Emerg Infect Dis 14: 349-350.
- Ulett KB, Benjamin WH Jr, Zhuo F, Xiao M, Kong F, Gilbert GL, Schembri MA, Ulett GC 2009. Diversity of group B streptococcus serotypes causing urinary tract infections in adults. J Clin Microbiol 47: 2055-2060.
Publication Dates
-
Publication in this collection
19 Apr 2011 -
Date of issue
Mar 2011
History
-
Accepted
25 Jan 2011 -
Received
11 June 2010