Abstracts
Microorganisms for biological control are capable of producing active compounds that inhibit the development of phytopathogens, constituting a promising tool toob tain active principles that could replace synthetic pesticides. This study evaluatedtheability of severalpotentialbiocontrol microorganismsto produce active extracellular metabolites. In vitro antagonistic capability of 50 bacterial isolates from rhizospheric soils of "criolla" potato (Solanum phureja) was tested through dual culture in this plant with different plant pathogenic fungi and bacteria. Isolates that showed significantly higher antagonistic activity were fermented in liquid media and crude extracts from the supernatants had their biological activities assessed by optical density techniques. Inhibitory effecton tested pathogens was observed for concentrations between 0.5% and 1% of crude extracts. There was a correlation between the antimicrobial activity of extracts and the use of nutrient-rich media in bacteria fermentation. Using a bioguided method, a peptidic compound, active against Fusarium oxysporum, was obtained from the 7ANT04 strain (Pyrobaculum sp.). Analysis by nuclear magnetic resonance and liquid chromatography coupled to mass detector evidenced an 11-amino acid compound. Bioinformatic software using raw mass data confirmed the presence of a cyclic peptide conformed by 11 mostly non-standard amino acids.
Solanum phureja; antagonistic bactéria; antibiosis; Fusarium oxysporum; bioactive metabolites
Os microorganismos biocontroladores têm capacidade de produzir compostos ativos que inibem o desenvolvimento de fitopatógenos, constituindo-se em uma ferramenta promissora para a obtenção de princípios ativos que substituem os pesticidas de sínteses. No presente trabalho, avaliou-se a capacidade de um conjunto de gentes com potencial biocontrolador, para produzir metabólitos extracelulares ativos. Avaliou-se a atividade antagônica in vitro de 50 isolamentos bacterianos provenientes de solos rizosféricos de batata (Solanum phureja) através de confrontamentos duplos com diferentes fungos e bactérias fitopatógenicas desta planta. Os isolamentos que mostraram atividade antagônica significativamente superior foram submetidos a fermentações líquidas e a partir dos sobrenadantes se obtiveram extratos brutos, cujas atividades biológicas foram determinadas empregando-se técnicas de densidade óptica. Evidenciou-se inibição sob os fitopatógenos avaliados para concentrações entre 0,5% para 1% dos extratos brutos. Observou-se uma correlação da atividade antimicrobiana dos extratos com o emprego de meios de cultivo ricos em nutrientes nas fermentações bacterianas. De forma biodirigida, isolou-se um composto de natureza peptídica com atividade contra Fusarium oxyporum a partir da cepa 7ANT04 (Pyrobaculum sp.). Com base nas análises por ressonância magnética nuclear e cromatografia líquida acoplada a detector de massas evidenciou-se um composto constituído por onze aminoácidos. O processamento dos dados de massas, mediante softwares de bioinformática, confirmou que o composto isolado é um peptídeo cíclico constituído por onze aminoácidos, em sua maioria, não comuns.
Solanum phureja; bactérias antagonistas; antibiosis; Fusarium oxysporum; metabólitos bioactivos
ARTIGOS
Antimicrobial activity of extracellular metabolites from antagonistic bacteria isolated from potato (Solanum phureja) crops
Atividade antimicrobiana de metabólitos extracelulares das bactérias antagonistas isoladas de cultivos de batata (Solanum phureja)
Sinar David Granada GarcíaI,III,* Autor para correspondência: Sinar David Granada García ( davidgranada888@gmail.com) ; Antoni Rueda LorzaII; Carlos Alberto PeláezIII
ICorporación para Investigaciones Biológicas (CIB), Unidad de Fitosanidad y Control Biológico. Carrera 72A No 78B 141. Medellín, Colombia
IIGrupo de Investigación en Productos Naturales (GIPRONUT), Universidad del Tolima. Barrio Santa Elena Parte Alta. Ibagué, Tolima
IIIGrupo Interdisciplinario de Estudios Moleculares (GIEM), Universidad de Antioquia. Calle 67 No 53-108. Medellín, Colombia
ABSTRACT
Microorganisms for biological control are capable of producing active compounds that inhibit the development of phytopathogens, constituting a promising tool toob tain active principles that could replace synthetic pesticides. This study evaluatedtheability of severalpotentialbiocontrol microorganismsto produce active extracellular metabolites. In vitro antagonistic capability of 50 bacterial isolates from rhizospheric soils of "criolla" potato (Solanum phureja) was tested through dual culture in this plant with different plant pathogenic fungi and bacteria. Isolates that showed significantly higher antagonistic activity were fermented in liquid media and crude extracts from the supernatants had their biological activities assessed by optical density techniques. Inhibitory effecton tested pathogens was observed for concentrations between 0.5% and 1% of crude extracts. There was a correlation between the antimicrobial activity of extracts and the use of nutrient-rich media in bacteria fermentation. Using a bioguided method, a peptidic compound, active against Fusarium oxysporum, was obtained from the 7ANT04 strain (Pyrobaculum sp.). Analysis by nuclear magnetic resonance and liquid chromatography coupled to mass detector evidenced an 11-amino acid compound. Bioinformatic software using raw mass data confirmed the presence of a cyclic peptide conformed by 11 mostly non-standard amino acids.
Additional keywords:Solanum phureja, antagonistic bactéria, antibiosis, Fusarium oxysporum, bioactive metabolites.
RESUMO
Os microorganismos biocontroladores têm capacidade de produzir compostos ativos que inibem o desenvolvimento de fitopatógenos, constituindo-se em uma ferramenta promissora para a obtenção de princípios ativos que substituem os pesticidas de sínteses. No presente trabalho, avaliou-se a capacidade de um conjunto de gentes com potencial biocontrolador, para produzir metabólitos extracelulares ativos. Avaliou-se a atividade antagônica in vitro de 50 isolamentos bacterianos provenientes de solos rizosféricos de batata (Solanum phureja) através de confrontamentos duplos com diferentes fungos e bactérias fitopatógenicas desta planta. Os isolamentos que mostraram atividade antagônica significativamente superior foram submetidos a fermentações líquidas e a partir dos sobrenadantes se obtiveram extratos brutos, cujas atividades biológicas foram determinadas empregando-se técnicas de densidade óptica. Evidenciou-se inibição sob os fitopatógenos avaliados para concentrações entre 0,5% para 1% dos extratos brutos. Observou-se uma correlação da atividade antimicrobiana dos extratos com o emprego de meios de cultivo ricos em nutrientes nas fermentações bacterianas. De forma biodirigida, isolou-se um composto de natureza peptídica com atividade contra Fusarium oxyporum a partir da cepa 7ANT04 (Pyrobaculum sp.). Com base nas análises por ressonância magnética nuclear e cromatografia líquida acoplada a detector de massas evidenciou-se um composto constituído por onze aminoácidos. O processamento dos dados de massas, mediante softwares de bioinformática, confirmou que o composto isolado é um peptídeo cíclico constituído por onze aminoácidos, em sua maioria, não comuns.
Palavras-chave adicionais:Solanum phureja, bactérias antagonistas, antibiosis, Fusarium oxysporum, metabólitos bioactivos.
Research on compounds derived from microbial sources has been of great importance due to the broad structural diversity and the variety of biological activities reported for this type of molecules (1), such as antitumor (1), antiviral (18) and antimicrobial (antibacterial, antifungal, antiprotozoal (1,8)) activities. Thus, many of the studies in the last decade have focused on isolating and evaluating compounds in different interactions in nature involving microorganisms. The challenges to be faced currently are the emergence of resistant pathogens, a decrease in the discovery of new active principles and legal requirements to use substances that generate a lower impact on the environment.
There are antagonistic interactions between microorganisms in which one of them limits the development of the other by various biochemical mechanisms. This capacity of some microbes makes them viable for use in biological control of plant pathogens (7). Biocontrol agents are not only an alternative to the intensive use of agrochemicals, but also happen to be a potential source of active principles that could replace, partially or totally, synthetic compounds. In addition, compared to other sources of biologically active molecules such as plants and animals, microorganisms stand out due to the possibility of obtaining a satisfactory scaling for mass culture through fermentation in bioreactors (6).
The present study evaluated the activity of metabolites obtained from fermentation of bacteria with antagonistic capacity against phytopathogens. The biosynthesis of active extracellular compounds produced by the antagonist was correlated with the observed inhibition. Moreover, physiological aspects that influence the production of these substances were studied, particularly a peptidic molecule obtained by bioguided isolation from one of the active extracts.
MATERIALS AND METHODS
Microorganisms
Fifty potentially antagonistic bacterial isolates obtained from rhizospheric soil of "criolla" potato (Solanum phureja) were used. These isolates were provided by the Agricultural Microbiology Group of the Biotechnology Institute at Universidad Nacional de Colombia (IBUN). Isolates were maintained and replicated in agar and Luria Bertani (LB) broth (10 g L-1 tryptone, 5 g L-1 yeast extract, 10 g L-1 NaCl, pH 7) at 30 ºC. Eight pathogens of importance in potato production system were employed: Rhizoctonia solani, Fusarium oxysporum ATCC 15648, Colletotrichum gloeosporioides ATCC 58696 and Alternaria sp., grown on potato dextrose agar (PDA Merck ®, Germany) at 25 ºC; and Ralstonia solanacearum, Pectobacterium carotovorum, Pseudomonas syringae and Xanthomonas campestris, grown in LB at 30 ºC. Some of these phytopathogens belong to the culture collection at the Plant Health and Biological Control Unit of CIB, while others were provided by Dr. Silvia Restrepo at Universidad de los Andes.
Preparation of inocula and in vitro antagonism assays
Inocula of bacteria, both antagonistic and plant-pathogenic, were prepared by transferring a loopful from a Petri dish into LB broth tubes and the concentration was adjusted according to the McFarland scale (pattern 0.5). For plant pathogenic fungi, with the exception of R. solani, suspensions of conidia were prepared from Petri dishes cultivated during 8 to 16 days in PDA at 25ºC. The obtained suspension was passed through a 70-µm filter (BD FalconTM, USA) and the concentration was determined by using a hemocytometer. Regarding R. solani, mycelium was fragmented by employing sterile glass beads and vortexed for one minute. Subsequently, filtration (70 µm) was held and the concentration was determined by counts of hyphae as colony forming units. For antagonism assays against plant pathogenic bacteria, these were massively seeded on LB agar, spreading 100 µL of the suspension onto the surface. Each Petri dish was divided into four sections and a 6.3mm-diameter Sensi-Disc (Schleider and Schuell Inc., England) was placed into each quadrant at 1 cm from the edge of the dish. An inhibition control was applied on one of the discs, consisting of 20 μL of chloramphenicol and streptomycin solution at concentrations of 50 and 1000 μg mL-1, respectively. On each of the three remaining discs, 20 μL of a suspension of one of the potentially antagonistic bacteria were added. A different bacterial isolate was applied on each Sensi-Disc and the zone of inhibition was measured after 24 h of incubation at 30 ºC. Growth controls were prepared by using bacteria alone and all the procedures were performed in quadruplicate and repeated once. Assays against plant pathogenic fungi were similarly done. PDA and a 1 x 104 conidia mL-1 suspension were used in this case. For R. solani, rather than suspension, a sclerotium was seeded on the center of the Petri dish. Inhibition control consisted of 20 μL of a Voriconazole, Cycloheximide and Ketoconazole solution at 1.25 and 10 μg mL-1, respectively; and the zone of inhibition was measured after 72 h of incubation at 25 ºC.
Antagonist growth curves
Bacterial inoculum (10 mL) was prepared in LB broth at a concentration of 0.5 McFarland and transferred to an Erlenmeyer flask with 90 mL of the same medium. Cultures were incubated on an orbital shaker at 30 ºC and 150 rpm. Aliquots (1 mL) were taken in quadruplicate and measured at 600 nm in a spectrophotometer from 0 to 72 h. The absorbance target was adjusted with uninoculated sterile LB medium.
Fermentation and extraction of 1 L of broth
An inoculum of bacterium to be fermented was prepared in 10 mL of broth, adjusting the concentration to 0.5 McFarland. It was taken to a total volume of 1 L and incubated for 96 h on an orbital shaker at 30 ºC and 150 rpm. To obtain extracts, biomass was separated from the fermented broth by using a tangential filter of 0.45 µm (Millipore Corporation USA). The supernatant was frozen and freeze-dried. A solid-liquid extraction was applied to the retrieved solid by using three 150 mL portions of methanol (Merck®, Germany). The solvent was removed by vacuum-assisted rotary evaporation, resulting in the crude extract.
Evaluation of the antimicrobial activity of extracts
The biological activity of extracts obtained from isolates 4ANT08, 7ANT04 and 7ANT05 and fractions obtained from 7ANT04 were assessed through optical density measurements at 595 nm on 96-well microplates, using a BIO-RAD plate reader model 680 XR. Sabouraud broth (Becton Dickinson®, USA) and water, both autoclave-sterilized, and aqueous solutions from treatments, sterilized for 25 minutes under ultraviolet light, were employed. Treatment effect was assessed by comparing the growth rate of treated fungus with respect to control. Medium (100 µL), treatment (50 µL) and inoculum (50 µL) of the fungus being evaluated (1 x 104 UFC mL-1) were added to each treated well. In addition, a growth control of the untreated plant pathogen and a contamination control, containing culture medium and sterile water, were used. Eight replicates of each well were performed. The plate was incubated at 25 ºC and its optical density measured at 595 nm, including previous agitation, at every 24 h. The percentage of inhibition was calculated by using the expression:
Inhibition (%) = [(control growth rate treatment growth rate) / control growth rate] x 100%
Effect of culture medium and fermentation time on antimicrobial activity
The effect of culture medium on the evaluated extracts was determined by using minimal medium (200 mL M9: 64 g L-1 Na2HPO4.7H2O, 15 g L-1 KH2PO4, 2.5 g L-1 NaCl, 5 g L-1 NH4Cl; 2 mL 1 M MgSO4; 25 mL glucose 20%; 0.1 mL 1 M CaCl2; 773 mL water), nutrient broth (5 g L-1 peptone, 3 g L-1 meat extract), 0.1% peptone broth (1 g L-1 peptone, 7 g L-1 NaCl), LB broth and modified TSB (1 g L-1 meat extract, 5 g L-1 glucose, 30 g L-1 broth (Merck®, Germany)). Test tubes with 5 mL of each medium in duplicate were inoculated with a loopful of the antagonistic bacteria and incubated for 96 h at 30 ºC and 180 rpm. Subsequently, each tube was subjected to centrifugation for 5 min at 7000 rpm, 25 ºC. The supernatant was frozen, freeze-dried, and extracted with two 2 mL portions of methanol. The solvent was removed by vacuum-assisted rotary evaporation. For time effect evaluation, fermentation of 1 L of broth was done, as indicated above, using modified TSB. Two 5 mL aliquots of fermentation were taken at different times (0, 24, 48, 72 and 96 h) and subjected to the same process of centrifugation, freeze-drying and extraction, as described above.
Chromatographic analysis and fractionation of extracts
High performance liquid chromatography (HPLC) was employed for analyses by using an Agilent 1200 Series (Agilent technologies, Germany) with UV-DAD detection (UV with diode array). Fractionation of the extract obtained from 7ANT04 isolate was carried out according to the results of antifungal activity. Fractionation was performed by preparative HPLC with prior scaling in analytical mode. Samples to be fractionated were dissolved in the proper proportion of methanol:water (HPLC grade methanol, Merck®, Germany; Mili-Q water) and passed through a 0.45-µm filter. An UltrasphereTM C18 column (150 x 2 mm, 5 µm) (Beckman, USA) was used for analytical runs and an Eclipse XDB-C18 (250 x 21.6 mm, 7 µm) (Agilent, USA) for the preparative scale. The solvent flow used in the analytical run was of 0.5 mL min-1 and of 30 mL min-1 in the preparative scale. The scaling of the sample injection volume from the analytical to the preparative method was performed by using the expression:
Preparative volume = analytical volume x (preparative column radius2 / analytical column radius2) x (preparative column length / analytical column length)
In all cases, mobile phases were used with different proportions of methanol:water until the best peak resolution was obtained.
Spectroscopic analysis and bioinformatics
Processing of samples for the spectroscopic analyses of compounds was developed at the Biochemistry and Plant Biotechnology Laboratory and the Organic Synthesis Group at the Universidad de Jaume I in Castellon, Spain. Experiments of nuclear magnetic resonance (NMR) were performed in a 500 MHz Varian spectrometer (Varian Inc., USA). Chemical shifts (δ) were expressed as ppm and coupling constants as Hertz (J). Center of peaks of the deuterated pyridine was used as an internal reference. The sample was dissolved in deuterated methanol. 1H NMR spectra were recorded at 500 MHz (1H frequency) in CD3OD solution at 30 ºC. 13C NMR signal multiplicity was determined with the DEPT sequence pulse at 125 MHz. HPLC-Mass spectrometry (HPLC-MS) analyses were determined by using an Agilent 1200 series via direct infusion, with positive mode electrospray ionization (ESI) and time of flight (TOF) detector (Agilent technologies, Germany). The sample was prepared at a concentration of 500 µg mL-1 in methanol:acetic acid 99:1. ESI conditions were: capillary voltage, 200 V; supply voltage, 4.5 kV; capillary temperature, 300ºC; nebulizer pressure, 15 psi, and gas flow 5 L min-1.
PEAKS 5.3 (10) and NRP-Tagging (11) were employed for bioinformatic analyses. Raw data obtained from the Agilet software was converted from ".yep" to ".mzXLM" using CompassXport (available on http://ionsource.com/functional_reviews/CompassXport/CompassXport.htm). Regarding NRP-Tagging, data in ".mzXML" format were converted to ".dta" with the help of PEAKS 5.3.
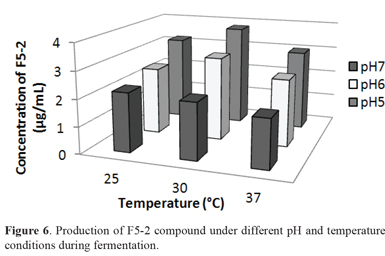
F5-2 production analysis under different pH and temperature conditions during fermentation
Tube fermentation (5 mL) was conducted in triplicate, using a 3 x 3 factorial arrangement (three incubation temperatures: 25, 30 and 37 ºC, and three different pH values: 5, 6 and 7) in a completely randomized design. An HPLC method was developed for the F5-2 compound quantification, consisting of a gradient from 60:40 methanol:water to 100% methanol from 0 to 10 min, followed by 5 additional minutes in 100% methanol. The calibration curve was built from solutions of the pure compound with concentrations between 1 and 20 µg mL-1 and read at 225 nm. Standards were run in triplicate and the calibration curve was calculated by linear regression. Compound quantification in fermentation supernatants was carried out by direct injection of a 20 µL aliquot.
RESULTS AND DISCUSSION
In vitro antagonism assays
Of the 50 isolates evaluated as potential antagonists, 18 showed antagonistic activity against at least one of the pathogens. Xanthomonas campestris and Alternaria sp. were the most susceptible strains (Table 1), whereas Pectobacterium carotovorum and Pseudomonas syringae did not show inhibition halos against any of the 50 isolates (data not shown). Another important fact was the resistance shown by Fusarium oxysporum against the evaluated isolates, with the exception of 7ANT04 (figure 1). Results show potential for production of active compounds from the isolates 4ANT08, 7ANT04 and 7ANT05, not only as a result of presenting the greatest inhibition halos, but also due to their spectrum of activity against several of the tested fungi, including X. campestris bacterium, in the case of 7ANT04.
Extraction of the most promising isolates and evaluation of their antifungal activity
After extraction of supernatants from the fermentation of 4ANT08, 7ANT04 and 7ANT05, yields between 4 and 6 g of crude extract per liter of fermented broth were obtained. Starting from these extracts, aqueous solutions were prepared and their inhibitory ability against R. solani, F. oxysporum and C. gloeosporioides was tested, respectively. Controls evidenced an increased optical density over time, whereas treatments showed no significant change. At the evaluated concentration (1%), crude extracts of 4ANT08 and 7ANT04 showed 100% inhibition, whereas for 7ANT05 extract it was 93%. This indicates that it was possible to extract some compounds, possibly involved in the antagonistic activity, by solid-liquid extraction with methanol from freeze-dried supernatants (9). However, there are reports where supernatants of fermentation without extraction have produced significant inhibition of the tested pathogens (10).
Effect of culture medium and fermentation time
The activity of crude extracts obtained from fermentation of antagonistic bacteria in different culture media was evaluated to determine which of them was the most appropriate for production of active compounds. These assays were carried out at a 0.5% concentration of extract. In the case of 4ANT08, it is evident that, with the exception of the nutrient broth, all evaluated media exhibit biological activity. This indicates that for this isolate there was no marked effect of the environment on the production of active metabolites (Figure 2-I). The two remaining isolates revealed greater fermentation inhibition by extracts in modified TSB and minimum media (Figure 2, II and III). This suggests that the tested antagonists do not have substantial nutritional requirements, since using a source of carbon and nitrogen, either organic or inorganic, allows them to develop in a normal way and at the same time exploit their metabolic potential.
Time effect was evaluated by following kinetics of the biological activity of aliquots taken from bacterial fermentation in modified TSB. This evaluation was performed only on isolates 4ANT08 and 7ANT04, since they showed a greater potential in the production of active metabolites in comparison with 7ANT05. In this case, the evaluated concentration was 0.5%. The highest inhibitory capacity was observed for the 96 h fermentation extract, both for 7ANT04 and 4ANT08 (Figure 3, I and II). However, it is clear that the activity begins to be significant after 24 h, moment in which the stationary phase is reached, as determined in the antagonist growth curves (data not shown). HPLC profiles of 7ANT04 extracts were obtained at different times. These profiles are shown in Figure 4.
Bioguided fractionation
After determining the best conditions for the antagonist in the production of active metabolites, four 1-L scale fermentation of 7ANT04 isolate were carried out by using modified TSB and a 96 h fermentation time. After the freeze-drying and extraction process of the supernatant, the obtained 28 g of extract were subjected to a bioguided fractionation. A 10-min gradient from 0 to 100% methanol was initially used, followed by 5 min in 100% methanol. Six fractions were collected (F1 to F6) and their activity was assessed at 0.125% after removing the solvent. Less polar fractions (F5 and F6) had the highest activity (Table 2). Additionally, these results correlate with those observed in chromatograms from the fermentation kinetics where an increase in less polar metabolites can be seen over time (Figure 3). Although F6 presented an inhibition percentage slightly higher than F5, its yield was low and it was not possible to continue with its fractionation.
Three compounds (F5-1, 2 and 3) were isolated from F5 and one of them was purified, the F5-2, which underwent a preliminary analysis in relation to the structural aspects of the molecule. Besides, dose-response assays were conducted with the compound in a range of concentrations between 30 and 500 µg mL-1, showing a minimum inhibitory concentration of 250 µg mL-1 (Table 3). A logarithmic correlation was found between compound concentration and its activity in the range from 250 to 30 µg mL-1 (R2 = 0.9978, p = 0.0000).
Structural characteristics of the F5-2 compound
13 C NMR signals (125 MHz, CD3OD)
Carbonylic carbons: δ 176.24 (CO), 173.87 (CO), 173.82 (CO), 173.38 (CO), 173.29 (CO), 173.01 (CO), 172.78 (CO), 172.52 (CO), 171.64 (CO), 171.46 (CO), 171.37 (CO).
Arylic carbons: 155.93 (C-OH, Ar), 129.76 (CH, Ar), 127.51 (CH, Ar), 115.02 (CH, Ar)
α Carbons: 61.87 (CH), 61.43 (CH), 56.36 (CH), 56.01 (CH), 51.10 (CH), 50.40 (CH), 50.17 (CH), 46.60 (CH).
R Group Carbons of different amino acids: 42.27 (CH), 38.84 (CH2), 36.40 (CH2), 36.35 (CH2), 35.34 (CH2), 35.13 (CH2), 34.91 (CH2), 34.85 (CH2), 34.28 (CH2), 30.37 (CH2), 29.73 (CH2), 29.65 (CH2), 29.42 (CH2), 29.35 (CH2), 29.32 (CH2), 29.16 (CH2), 29.06 (CH2), 28.82 (CH2), 27.72 (CH2), 27.13 (CH2), 26.82 (CH2), 26.34 (CH3), 25.58 (CH3), 24.79 (CH3), 21.60 (CH3), 18.20 (CH3), 10.30 (CH3).
1 H NMR signals (500 MHz, CD3OD).
Amide protons: δ 8.54 (s), 8.36 (d, J = 5.6 Hz), 8.23 (d, J = 7.6 Hz), 8.08 (d, J = 7.4 Hz), 7.96 (d, J = 8.1 Hz), 7.93 (d, J = 5.4 Hz), 7.71 (d, J = 7.9 Hz), 7.57 (s), 7.46 (d, J = 19.0 Hz), 7.40 - 7.36 (m), 7.36 - 7.31 (m).
Arylic protons: δ 7.06 (d, J = 8.1 Hz, 2H), 6.73 (d, J = 8.3 Hz, 2H)
αC-bound protons: 4.68 (dd, J = 9.8, 4.7 Hz, 1H), 4.62 (t, J = 4,2 Hz, 1H), 4.59 (t, J = 4,2 Hz, 1H), 4.51 (t, J = 5.9 Hz, 1H), 4.41 (t, J = 4.4 Hz, 1H), 4.30 (dd, J = 9.4, 4.7 Hz, 1H), 4.22 (t, J = 7.3 Hz, 1H), 4.17 (dd, J = 10.1, 4.0 Hz, 1H), 4.03 (dd, J = 11.6, 5.1 Hz, 1H), 3.95 (dt, J = 10.8, 6.2 Hz, 1H), 3.84 3.75 (m, 2H)
R Group protons of different amino acids:3.34 (s, 1H), 3.11 (dd, J = 14.4, 4.6 Hz, 1H), 2.95 2.83 (m, 3H), 2.76 (dd, J = 15.4, 8.5 Hz, 1H), 2.67 (dd, J = 15.4, 4.9 Hz, 1H), 2.61 2.44 (m, 3H), 2.40 (d, J = 15.5 Hz, 1H), 2.31 (t, J = 7.0 Hz, 3H), 2.26 (dt, J = 12.5, 7.1 Hz, 2H), 2.22 2.07 (m, 2H), 1.99 (dddd, J = 24.6, 19.6, 11.8, 6.8 Hz, 3H), 1.63 (dt, J = 12.7, 6.9 Hz, 1H), 1.50 (tt, J = 14.8, 7.0 Hz, 2H), 1.39 1.22 (m, 17H), 1.16 (dt, J = 12.8, 6.0 Hz, 4H), 0.91 0.83 (m, 6H).
The spectroscopic features reported by Wishart et al. (16, 17), in addition to the obtained chromatographic information, are the basis for the hypothesis that the F5-2 compound is a cyclic peptide consisting of eleven amino acid residues in which R groups do not have easily ionizable functional groups. Information obtained by chromatography consisted in the use of mobile phases employing buffer solutions at different pH (2.5, 7 and 9) where retention time of the F5-2 compound remained unchanged. This indicates not only that the R groups of the amino acids do not possess ionizable functional groups, but also that this peptide does not have the carbonyl and amino terminal exposed as in the linear peptides, which shows that it is cyclical. A [M + H] with m/z of 1065.6 was detected by HPLC-MS, indicating a molecular weight of 1064.6 Da. Regarding the compound spectroscopic information, eleven non-ketone carbonylic carbons are observed in the 13C NMR spectrum, which show displacement towards higher field, suggesting a link with a nitrogen atom. Similarly, the proton spectrum confirms the presence of eleven amide protons in the characteristic range, despite changes generated by the solvent and the variability of this type of proton (16, 17). In the displacement range between 46 and 62 ppm, eight methine signals characteristic of the common amino acids αC can be found (2, 16). Although eleven αC were expected, some of these signals could have overlapped with the solvent signal (48-50 ppm). There are also carbons belonging to an aromatic system, two of them quaternaries; at the same time, one of them is bound to an OH, and four methines, two of them are equivalent due to the para-substitution. This fact is confirmed in the proton spectrum where two coupled signals with an average constant of 8.2 Hz are observed and integrated to two protons. Signals corresponding to methylenes and methyl groups, characteristic of the R group of common amino acids, were found at the aliphatic region of the carbon spectrum (2, 15).
As observed in the aromatic region of the spectra, it is possible to say that one of the amino acids has high correspondence with a tyrosine residue.Moreover, high absorbance of the compound in the ultraviolet is due to this tyrosine, observing a typical UV spectrum with a maximum at 276 nm and a shoulder at 225 nm.Also, it can be concluded that the molecule has a glycine residue caused by the αC signal in 46.60 ppm and the signal between 3.84 and 3.75 ppm in the 1H NMR spectrum corresponding to the diastereotopic protons of this amino acid. It is possible to obtaininformation from the observed fractionation in the mass spectrum, where typical losses corresponding to amino acids such as leucine, isoleucine, alanine and serine can be found. However, due to the atypical fractionation of the cyclic peptides, it is not easy to determine the sequence of amino acids forming the molecule. Similarly, obtaining appropriate two-dimensional spectra that could provide evidence for the connectivity of amino acids was not possible.
Bioinformatic analysis using PEAKS 5.3 and NRP-Tagging
In the case of PEAKS 5.3 analysis, the results showed an average local confidence (ALC) of 51% for the AGPNYNGCVNG sequence, being the only one coinciding with a molecular ion of 1065.5 Da. Nevertheless, a result to be noted is the nature of the amino acids given by PEAKS that only shows neutral amino acids and the presence of a tyrosine, which is consistent with that observed in the NMR and the HPLC.
Results of NRP-Tagging analysis generated homologies with cyclic sequences from six to eleven fragments. The larger number of homologies were found for sequences of nine fragments (18 sequences) and ten fragments (19 sequences); however, the highest homology value (86.20%) was found for a cyclic sequence of eleven fragments with masses of 87-62-111-97-114-59-69-149-74-58-184 Da, but it is only possible to assign identity to subunits 87, 97 and 114, corresponding to serine, proline and asparagine, respectively. Among sequences of ten fragments, sequence 87-62-111-97-114-59-218-74-58-184 showed an 85.10% homology, while the sequence of nine fragments with higher homology (83.99%) was 87-62-111-97-114-59-292-58-184. All these cyclic sequences have eight fragments in common (87-62-111-97-114-59 - ... -58 to 184). That is, the most significant inconsistencies found by the NRP-Tagging algorithms occur in a fragment of 292 Da. Finally, it can be deduced through this tool that most of the fragments or amino acid residues forming the structure have some type of modification or are uncommon amino acids. This proves to be a valuable tool, but it needs to be complemented with spectra information from 2D NMR. A model for the structure of the F5-2 molecule is shown in Figure 5.
Production of F5-2 under different conditions of pH and temperature during fermentation
The F5-2 production at different pH and temperature conditions showed significant differences (p <0.05), with a maximum of 3.8 ± 0.5 μg mL-1 at pH 5 and 30 ºC, and a minimum of 1.8 ± 0.4 μg mL-1 at pH 7 and 37 ºC (Figure 6). These results reveal a potential use for the 7ANT04 bacterium as a crop protection inoculum, since reported conditions for the highest crop productivity (soil pH between 5-5.5) (14) are similar to the conditions wherein the microorganism exhibited its maximum productivity.
Thirty-six percent of the 50 tested bacterial isolates, obtained from soils cultivated with "criolla" potato (Solanum phureja), showed antagonistic activity against crop plant pathogens. These are a source not only of biocontrol microorganisms, but also of bioactive substances with potential use for controlling plant pathogens associated with potato cultivation.
The antagonism and extraction of metabolic product assays, performed out of fermentation of antagonistic bacteria in liquid medium, highlight the better antifungal activity of 7ANT04 isolate extracts against F. oxysporum. The potential of this isolate is not only seen through antagonism assays, but also by evaluating its crude extracts at a concentration of 0.5%. Isolates 7ANT04 and 4ANT08 were identified by the Agricultural Microbiology Group of IBUN as Pyrobaculum sp. and Pseudomonas sp., respectively. The genus Pseudomonas has been widely reported as an antagonist of a variety of plant pathogens (5, 12), but no reports are known for Pyrobaculum as antagonistic microorganism, to our knowledge. Selection of these two isolates, amid 18 that showed antagonistic capacity, was based on their activity spectrum and the high inhibitory capacity of their extracts.
Isolate 7ANT05, despite the broad spectrum exhibited in the antagonism assays, showed low inhibition compared to the extracts of 4ANT08 and 7ANT04 isolates which completely inhibited the growth of pathogens at the same concentration (0.5%).
We found an effect of culture medium during fermentation of the three isolates on the production of metabolites with activity against the evaluated plant pathogens. The greatest inhibition occurred during fermentation with minimal medium and modified TSB. This suggests that these bacteria have the ability to metabolize substrates both organic (TSB) and inorganic (MM). These two growth media have glucose content as a common characteristic, a differential factor from the other three evaluated media (NB, PB 0.1% and LB). Despite the fact that several studies reported glucose as catabolic suppressor of active secondary metabolite-producing microorganisms (4), our findings suggest glucose as a carbon source suitable for the production of active metabolites, which is consistent with that described by Saha et al. (13). On the other hand, it is important to evaluate other nutritional sources of carbon, and even nitrogen, which could maximize the production of active compounds by the antagonist.
The kinetics of biological activity in fermentation evidences production of active compounds when reaching the stationary phase of growth of the antagonistic microorganism. This suggests that a significant production of the active compounds is not reached until bacterium growth rate begins to decline, which is in agreement with that reported by Clark et al. (3). The fractionation of extracts showed a higher activity in fractions with less polar character, which correlates with the chromatographic profiles from the kinetics extracts where an increase in peaks found at higher retention times (10.5 to 12 min) can be recognized, and it is practically not observed in the extract profile at time zero (Figure 4). Furthermore, the relative abundance of these compounds is increased over time. However, there is no correlation between activity and time because the percentage inhibition after 24 h fluctuates until reaching a maximum value after 96 h (R2 = 0.508, p> 0.05). This suggests that not all the substances, responsible for the activity, accumulate as the fermentation progresses. Despite the structural complexity of the F5-2 molecule, other active molecules with a similar core were found having small variations in their NMR spectra. Therefore, the biological activity of the extract is not only attributed to the presence of this compound (F5-2), but to the combined effect among structurally related molecules.
ACKNOWLEDGEMENTS
Sustainability Program 2013-2014, University of Antioquia. Colombian Agriculture and Rural Development Minister.
Data de chegada: 22/11/2013.
Aceito para publicação em: 11/08/2014.
- 1. Bérdy, J. Bioactive microbial metabolites: a personal view. Journal of antibiotics, Budapest, v.58, n.1, p.126, 2005.
- 2. Arnold, M.R.; Kremer, W.; Ludemann, H.D.; Kalbitzer, H.R. 1H-NMR parameters of common amino acid residues measured in aqueous solutions of the linear tetrapeptides Gly-Gly-X-Ala at pressures between 0.1 and 200 MPa. Biophysical Chemistry, Regensburg, v.96, n.2-3, p.129-140, 2002.
- 3. Clark, G.J.; Bushell, M.E. Oxygen limitation can induce microbial secondary metabolite formation: investigations with miniature electrodes in shaker and bioreactor culture. Microbiology, Guildford, v.141, n.3, p.663-669, 1995.
- 4. Gallo, M.; Katz, E. Regulation of secondary metabolite biosynthesis: catabolite repression of phenoxazinone synthase and actinomycin formation by glucose. Journal of bacteriology, Washington, v.109, n.2, p.659-667, 1971.
- 5. Ganeshan, G.; Manoj, K. Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases. Journal of plant interactions, Bangalore, v.1, n.3, p.123134, 2005.
- 6. Huczyński, A.; Stefańska, J.; Przybylski, P.; Brzezinski, B.; Bartl, F. Synthesis and antimicrobial properties of monensin A esters. Bioorganic & medicinal chemistry letters, Grunwaldzka, v.18, n.8, p.2585-2589, 2008.
- 7. Köhl, J.; Postma, J.; Nicot, P.; Ruocco, M.; Blum, B. Stepwise screening of microorganisms for commercial use in biological control of plant-pathogenic fungi and bacteria. Biological control, Wageningen, v.57, n.1, p.1-12, 2011.
- 8. Krohn, K,; Biele, C.; Drogies, K-H.; Steingröver, K.; Aust, H-J.; Draeger, S.; Schulz, B. Fusidilactones, a New Group of Polycyclic Lactones from an Endophyte Fusidium sp. European journal of organic chemistry, Weinheim, v.14, n.1, p.2331-2336, 2002.
- 9. Li, Q.; Jiang, Y.; Ning, P.; Zheng, L.; Huang, J.; Li, G.; Jiang, D.; Hsiang, T. Suppression of Magnaporthe oryzae by culture filtrates of Streptomyces globisporus JK-1. Biological control, Wuhan, v.58, n.2, p.139-148, 2011.
- 10. Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid communications in mass spectrometry, London, v.17, n.20, p.2337-2342, 2003.
- 11. Ng, J.; Bandeira, N.; Liu, W.T.; Ghassemian, M.; Simmons, T.L.; Gerwick, W.H.; Linington, R.; et al. Dereplication and de novo sequencing of nonribosomal peptides. Nature methods, La Jolla, v.6, n.8, p.596599, 2009.
- 12. Páez, M.; Martínez-Nieto, P.; Bernal-Castillo, J. Siderophore producing Pseudomonas as pathogenic Rhizoctonia solani and Botrytis cinerea antagonists. Universitas Scientiarum, Bogotá, v.10, n.1, p.65-74, 2005.
- 13. Saha, M.; Ghosh, D.Jr.; Ghosh, D.; Garai, D.; Jaisankar, P.; Sarkar, K.K.; et al. Studies on the production and purification of an antimicrobial compound and taxonomy of the producer isolated from the marine environment of the Sundarbans. Applied microbiology biotechnology, Kolkata, v.66, n.5, p.497-505, 2005.
- 14. Srek, P.; Hejcman, M.; Kunzová, E. Multivariate analysis of relationship between potato (Solanum tuberosum L.) yield, amount of applied elements, their concentrations in tubers and uptake in a long-term fertilizer experiment. Field crop research, Prague, v.118, n.2, p.183-193, 2010.
- 15. Wishart, D.S.; Sykes, B.D. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. Journal of biomolecular NMR, Edmonton, v.4, n.2, p.171-80, 1994.
- 16. Wishart, DS. Interpreting protein chemical shift data. Progress in nuclear magnetic resonance spectroscopy, Edmonton, v.58, n.1-2, p.62-87, 2011.
- 17. Wishart, DS.; Bigam, C.; Holm, A.; Hodges, R.; Sykes, B. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor. Journal of biomolecular NMR, Edmonton, v.5, p.67-81, 1995.
- 18. Yu, B-Z.; Zhang, G-H,; Du, Z-Z.; Zheng, Y-T.; Xu, J-C.; Luo, X-D. Phomoeuphorbins A-D, azaphilones from the fungus Phomopsis euphorbiae Phytochemistry, Kunming, v.69, n.13, p.2523-2526, 2008.
Publication Dates
-
Publication in this collection
30 Oct 2014 -
Date of issue
Sept 2014
History
-
Accepted
11 Aug 2014 -
Received
22 Nov 2013