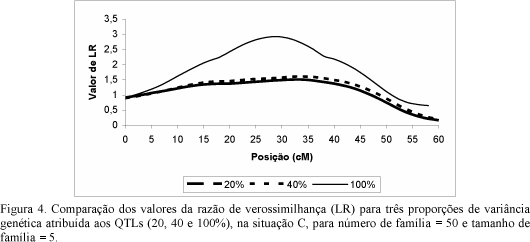
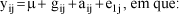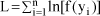A study was carried out by simulation to evaluate the efficiency and robustness of the random model approach for estimation of the QTL location and variance components in an outbred population with full-sib family structure. Two QTL were positioned in the chromosome in the same interval, in adjacent and at no adjacent intervals. The population was created with different sizes and numbers of families and variances due to QTL in a trait with h² = 0.25. The estimations of QTL parameters (locations and variance components) were based on the sib-pair approach. The proportions of genes identical-by-descent (IBD) at the two QTL were estimated from the IBD at two flanking markers. The most important factors afeccting the estimates of QTL parameters and power of detection were the proportion of variance due to QTL, and the number and size of the full-sibs families. Given a sufficient number of families and high proportions of genetic variance due to QTL, the random model approach can detect a QTL with high power and provides accurate estimates of the QTL position if there are no two QTL in the same interval.
genetic marker; interval mapping; random model; sib-pair method; QTL