ABSTRACT
BACKGROUND:
There has been little evidence to suggest that the IL-6 -174G>C and IL-10 -1082A>G polymorphisms are significantly associated with susceptibility to celiac disease. Thus, we performed the present meta-analysis to explore the potential association between these polymorphisms and celiac disease risk.
METHODS:
Eligible studies were searched in PubMed, Medline, Embase, Web of Science and CNKI database up to April 20, 2019. Odds ratios with 95% confidence interval were calculated to assess the potential associations. Moreover, we performed the heterogeneity, sensitivity, and publication bias tests to clarify and validate the pooled results.
RESULTS:
Overall, nine case-control studies involving five studies with 737 cases and 1,338 control on IL-6 -174G>C polymorphism and four studies with 923 cases and 864 controls on IL-10 -1082A>G polymorphism were selected. The pooled ORs showed that the IL-6 -174G>C and IL-10 -1082A>G polymorphisms were not significantly associated with increased risk of celiac disease under all five genetic models. There was no publication bias.
CONCLUSION:
To the best of our knowledge, this is the first meta-analysis summarizing all of the available studies on the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with celiac disease. Our results suggest that the IL-6 -174G>C and IL-10 -1082A>G polymorphisms may not be associated with increased risk of celiac disease. Moreover, large and well-designed studies are needed to fully describe the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with celiac disease.
HEADINGS:
Celiac disease; Interleukin-6; Interleukin-10; Genetic polymorphism
RESUMO
CONTEXTO:
Há poucas evidências para sugerir que os IL-6 -174G>C e IL-10 -1082A>G polimorfismos são significativamente associados com susceptibilidade para doença celíaca. Assim, foi realizada a presente meta-análise para explorar a potencial associação entre estes polimorfismos com o risco de doença celíaca.
MÉTODOS:
Foram pesquisados estudos elegíveis no Pubmed, Medline, Embase, Web of Science e CNKI Database até abril de 2019. Razões de probabilidades com 95% de intervalo de confiança foram calculados para avaliar as potenciais associações. Além disso, observou-se a heterogeneidade, a sensibilidade e o viés de publicação para esclarecer e validar os resultados agrupados.
RESULTADOS:
No total, nove estudos caso-controle envolvendo cinco estudos com 737 casos e 1.338 controle em IL-6 -174G>C polimorfismo e quatro estudos com 923 casos e 864 controles em IL-10 -1082A>G polimorfismo foram selecionados. As razões de probabilidade mostraram que o IL-6 -174G>C e IL-10 -1082A>G polimorfismos não estavam significativamente associados com aumento risco de doença celíaca nos cinco modelos genéticos. Não foi detectado viés de publicação.
CONCLUSÃO:
Pelo nosso conhecimento esta é a primeira meta-análise resumindo todos estudos disponíveis para associação de IL-6 -174G>C e IL-10 -1082A>G polimorfismos com doença celíaca. Estes resultados sugerem que os IL-6 -174G>C e IL-10 -1082A>G polimorfismos podem não ser associados com aumento risco de doença celíaca. Além disso, maiores estudos e mais bem desenhados são necessários para descrever totalmente a associação de IL-6 -174G>C e IL-10 -1082A>G polimorfismos com doença celíaca.
DESCRITORES:
Doença celíaca; Interleucina-6; Interleucina-10; Polimorfismo genético
INTRODUCTION
Celiac disease (CD), also called gluten intolerance, is a chronic and immune-mediated enteropathy with intestinal and extraintestinal manifestations triggered by the ingestion of gluten-containing grains in genetically predisposed individuals11. Rossi E, Basso D, Zambon C-F, Navaglia F, Greco E, Pelloso M, et al. TNFA Haplotype Genetic Testing Improves HLA in Estimating the Risk of Celiac Disease in Children. Goldberg AC, editor. PLoS One. 2015;10:e0123244.
2. Barartabar Z, Nikzamir A, Sirati-Sabet M, Aghamohammadi E, Chaleshi V, Rostami Nejad M, et al. The relationship between 174 G/C and -572 G/C of IL-6 gene polymorphisms and susceptibility of celiac disease in the Iranian population. Gastroenterol Rev. 2018;13:293-8.-33. Aflatoonian M, Moghimi M, Akbarian-Bafghi MJ, Morovati-Sharifabad M, Jarahzadeh MH, Neamatzadeh H. Association of TNF- α-308G>A polymorphism with susceptibility to celiac disease: a systematic review and meta-analysis. Arq Gastroenterol. 2019;56:88-94.. A review of celiac disease studies showed a biopsy-proven prevalence ranging from 0.15% to 1.9% in unselected populations of North America and Western Europe44. Dubé C, Rostom A, Sy R, Cranney A, Saloojee N, Garritty C, et al. The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology. 2005;128(4 Suppl 1):S57-67.. The diagnosis of CD can take several years to confirm and often times the diagnosis can be delayed by patients with a normal or high body mass index (BMI)55. Cheng J, Brar PS, Lee AR, Green PHR. Body Mass Index in Celiac Disease. J Clin Gastroenterol. 2010;44:267-71..
The actual etiology of CD is not fully understood, but some risk factors have been implicated in its development, including an association of genetic and environmental factors66. Capilla A, Donat E, Planelles D, Espinós C, Ribes-Koninckx C, Palau F. Genetic analyses of celiac disease in a Spanish population confirm association with CELIAC3 but not with CELIAC4. Tissue Antigens. 2007;70:324-9.,77. Khan S, Mandal RK, Jawed A, Dar SA, Wahid M, Panda AK, et al. TNF-α -308 G > A (rs1800629) Polymorphism is Associated with Celiac Disease: A Meta-analysis of 11 Case-Control Studies. Sci Rep. 2016;6:32677.. Nevertheless, few environmental factors such as Infant feeding practices that influence the risk of having CD have been identified88. Akobeng AK, Ramanan A V, Buchan I, Heller RF. Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies. Arch Dis Child. 2005;91:39-43.. The genetics of CD is closely related to HLA class II molecules HLA-DQ2 or HLA-DQ8, and has received attention for a decade with numerous studies published and several recent reviews. Moreover, genome-wide association studies (GWAS) have expanded our understanding of genetic predisposition and revealed contribution of genes encoding for the pro-inflammatory cytokines in susceptibility to CD99. Broce I, Karch CM, Wen N, Fan CC, Wang Y, Hong Tan C, et al. Immune-related genetic enrichment in frontotemporal dementia: An analysis of genome-wide association studies. Brayne C, editor. PLOS Med. 2018;15:e1002487..
The human IL-6 gene is mapped to chromosome 7p21-24, consists of seven exons and spanning 12.8 kb of genomic DNA1010. Ren H, Zhang Y, Yao Y, Guo T, Wang H, Mei H, et al. Association between the interleukin-6 genetic polymorphism 174 G/C and thrombosis disorder risk. Medicine (Baltimore). 2016;95:e4030.. Moreover, the IL-10 gene is mapped to the long arm of chromosome 1 (1q31-32), composed of five exons and spans about 4.7 kb1111. Namazi A, Forat-Yazdi M, Jafari M, Farahnak S, Nasiri R, Foroughi E, et al. Association of Interleukin-10 -1082 a/G (Rs1800896) Polymorphism With Susceptibility To Gastric Cancer: Meta-Analysis of 6,101 Cases and 8,557 Controls. Arq Gastroenterol . 2018;55:33-40.,1212. Sheikhpour E, Noorbakhsh P, Foroughi E, Farahnak S, Nasiri R, Neamatzadeh H. A survey on the role of interleukin-10 in breast cancer: A narrative. Reports Biochem Mol Biol. 2017;7(1).. The importance of IL-6 and IL-10 in many physiological and pathological processes, particularly in the autoimmune disease, has been reported1313. Hirano T. Interleukin 6 in autoimmune and inflammatory diseases: a personal memoir. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:717-30.,1414. Beebe AM, Cua DJ, de Waal Malefyt R. The role of interleukin-10 in autoimmune disease: systemic lupus erythematosus (SLE) and multiple sclerosis (MS). Cytokine Growth Factor Rev. 2002;13:403-12.. In recent decade, a number of studies have reported the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD susceptibility. However, those studies have drawn inconsistent conclusions, because individual study with small sample sizes may be underpowered to detect the association of L-6 -174G>C and IL-10 -1082A>G polymorphisms on susceptibility of CD. In order to get more precision results for the polymorphism and the risk of CD, we carried out a first meta-analysis including all eligible studies published to date to comprehensively examine the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with susceptibility to CD.
METHODS
Identification and eligibility of relevant studies
A comprehensive computer searches were carried out in PubMed, Google Scholar, EBSCO, EMBASE, Web of science, Scientific Information Database (SID), Islamic World Science Citation Center (ISC), Wanfang, Ovid, Weipu, China National Knowledge Infrastructure (CNKI) database to collect case-control studies related to the association of association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD up to April 20, 2019. The keywords were as follows: (‘’celiac disease’’ OR ‘’ coeliac disease’’ OR ‘’gluten intolerance’’ OR ‘’celiac sprue’’ OR ‘’nontropical sprue’’ OR ‘’endemic sprue’’) AND (‘’interleukin 6‘’ OR ‘’IL-6’’ OR ‘’-174G>C’’ OR ‘’rs1800795’’) AND (‘’ interleukin 10’’ OR ‘’IL-10’’ OR ‘’-1082A>G’’ OR ‘’rs1800896’’) AND (‘’SNPs’’ OR ‘’polymorphism’’ OR ‘’mutation’’ OR ‘’variation’’ OR ‘’allele’’). Furthermore, bibliographies of main retrieved case-control and review articles were also checked by a manual search to identify additional eligible studies. Searches were limited to published studies in Persian, Chinese and English.
Inclusion criteria and exclusion criteria
We selected eligible studies according to the following criteria: (1) studies with case-control and cohort design; (2) evaluated the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD; (3) adequate data to that odds ratios (ORs) with 95% confidence intervals (CIs); (4) the study had to be published using human subjects. Exclusion criteria were: (1) insufficient data on the distribution of IL-6 -174G>C and IL-10 -1082A>G loci genotypes; (2) case-only, linkage and family based studies; (3) case reports, posters, reviews, abstracts, letter to editors, previous meta-analyses; and (4) duplicates of previous studies or data. If a study was subsequently updated, we selected the study with the largest sample size. The authors independently checked all studies to examine whether the eligible studies fulfilled the inclusion criteria.
Data extraction
Two independent investigators reviewed and extracted data from all eligible publications based on the inclusion and exclusion criteria listed above. Discrepancies were adjudicated by a third author until consensus was reached on all items. The data extracted from each eligible study included the first author’s name, year of publication, number of cases and controls, ethnicity, country of study population, source or design of experiment (population- or hospital-based controls), genotyping methods, number of genotyped cases and controls for IL-6 -174G>C and IL-10 -1082A>G loci, Hardy-Weinberg equilibrium (HWE) in controls, and minor allele frequency (MAF). Disputes were settled by consulting the third person. The cases and controls ethnicities were categorized as Caucasian, Asian, African or Mixed.
Statistical analysis
In this meta-analysis, the crude odds ratio (OR) with the corresponding 95% confidence intervals (95% CI) were used to assess the strength of association of the IL-6 -174G>C and IL-10 -1082A>G polymorphisms with risk of CD. The Z-test was used to measure the significance of the pooled OR, and statistical significance was defined as P<0.05 (two-tailed). Five genetic models were used for pooling the ORs, i.e., allele (B vs. A), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB+BA vs. AA) and recessive (BB vs. BA+AA), respectively. The presence of heterogeneity between studies was evaluated with the Cochran’s Q statistic; in which P<0.10 indicated significant heterogeneity. Moreover, a Chi-square-based I2 test was used to quantify the degree of heterogeneity (I2<25%, no heterogeneity; 25%<I2<50%, moderate heterogeneity; 50%<I2<75%, large heterogeneity; I2>75%, extreme heterogeneity). Accordingly, the pooled ORs were examined using a fixed-effects (Mantel-Haenszel method) (if P>0.05 or I2<50%); otherwise, random-effects model (DerSimonian- Laird method) was utilized (if P<0.05 or I2>50%) based on the level of heterogeneity. For each case-control study, departure of the IL-6 -174G>C and IL-10 -1082A>G polymorphisms frequencies in control groups from the Hardy-Weinberg equilibrium (HWE) was examined using the chi-square test, and deviation was considered when P<0.05. We have tested our results stability and reliability using sensitivity analysis, in which one study was deleted each time and the analyses were repeated. Publication bias was tested with the funnel plot and Egger’s linear regression asymmetry test; P<0.05 suggested statistically significant publication bias. All analyses were performed with the Comprehensive Meta-Analysis (CMA) 2.0 software (Biostat, USA). Two-sided P-values <0.05 were considered statistically significant. If P value >0.05, the genotype distribution of the control group conformed to HWE.
RESULTS
Study characteristics
The selection process of eligible studies is shown in Figure 1. According to our search strategy, 93 articles were initially screened. After duplicates had been removed, then, 48 articles were determined to be irrelevant papers and not related to the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD and were initially excluded. Afterward, 19 studies that focused on other SNPs rather than IL-6 -174G>C and IL-10 -1082A>G polymorphisms, review articles, case only studies and studies with insufficient data polymorphism were excluded. Finally, a total of nine case-control studies involving five studies with 737 cases and 1,338 control on IL-6 -174G>C polymorphism1515. Garrote JA, Arranz E, Tellería JJ, Castro J, Calvo C, Blanco-Quirós A. TNF alpha and LT alpha gene polymorphisms as additional markers of celiac disease susceptibility in a DQ2-positive population. Immunogenetics. 2002;54:551-5.
16. Barisani D, Ceroni S, Meneveri R, Cesana BM, Bardella MT. IL-10 polymorphisms are associated with early-onset celiac disease and severe mucosal damage in patients of Caucasian origin. Genet Med. 2006;8:169-74.
17. Dema B, Martínez A, Fernández-Arquero M, Maluenda C, Polanco I, Figueredo MA, et al. The IL6-174G/C polymorphism is associated with celiac disease susceptibility in girls. Hum Immunol. 2009;70:191-4.
18. de Albuquerque Maranhão RM, Martins Esteves FA, Crovella S, Segat L, Eleutério Souza PR. Tumor necrosis factor-α and interleukin-6 gene polymorphism association with susceptibility to celiac disease in Italian patients. Genet Mol Res. 2015;14:16343-52.-1919. Barartabar Z, Nikzamir A, Sirati-Sabet M, Aghamohammadi E, Chaleshi V, Rostami Nejad M, et al. The relationship between 174 G/C and -572 G/C of IL-6 gene polymorphisms and susceptibility of celiac disease in the Iranian population. Gastroenterol Rev . 2018;13:293-8., and four studies with 923 cases and 864 controls on IL-10 -1082A>G polymorphism1616. Barisani D, Ceroni S, Meneveri R, Cesana BM, Bardella MT. IL-10 polymorphisms are associated with early-onset celiac disease and severe mucosal damage in patients of Caucasian origin. Genet Med. 2006;8:169-74.,2020. Hahn-Zoric M, Hytönen AM, Hanson LA, Nilsson LA, Padyukov L. Association of -1087 IL10 and -308 TNFA gene polymorphisms with serological markers of coeliac disease. J Clin Immunol. 2003;23:291-6.
21. Lio D, Scola L, Forte GI, Accomando S, Giacalone A, Crivello A, et al. TNFα, IFNγ and IL-10 gene polymorphisms in a sample of Sicilian patients with coeliac disease. Dig Liver Dis. 2005;37:756-60.-2222. Zupin L, Polesello V, Catamo E, Crovella S, Segat L. Interleukin-10 gene promoter polymorphisms in celiac patients from north-eastern Italy. Hum Immunol . 2014;75:656-61. were selected. The Characteristics of included studies are shown in Table 1. All of the selected papers were written in English and published between 2006 and 2018. Among them, eight studies were based on Caucasian (Spain, Italy, and Sweden), and one was based Middle-East Asian (Iran). The IL-6 -174G>C and IL-10 -1082A>G polymorphisms frequencies in each study, results of HWE test in control groups and MAFs are shown in Table 1. The distribution of genotypes in all studies was consistent with HWE except for two studies on c IL-6 -174G>C (Table 1).
Quantitative synthesis
IL-6 -174G>C polymorphism
Table 2 listed the main results of the meta-analysis of IL-6 -174G>C polymorphism with CD risk. When all the eligible studies were pooled into meta-analysis, the results showed that IL-6 -174G>C polymorphism was not significantly associated with increased risk of CD under all genetic models i.e., allele (A vs G: OR=0.984, 95% CI 0.857-1.131, P=0.820, Figure 2A), homozygote (AA vs GG: OR=1.101, 95% CI 0.804-1.507, P=0.548), heterozygote (AG vs GG: OR=0.654, 95% CI 0.378-1.311, P=0.128), dominant (AA+AG vs GG: OR=0.693, 95% CI 0.418-1.151, P=0.157), and recessive (AA vs AG+GG: OR=1.149, 95% CI 0.856-1.543, P=0.356).
Forest plot of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD. A) IL-6 -174G>C (allele model: A vs G); B) IL-10 -1082A>G (recessive model: AA vs AG+GG).
IL-10 -1082A>G polymorphism
Table 2 listed the main results of the meta-analysis of IL-10 -1082A>G polymorphism with CD risk. When all the eligible studies were pooled into meta-analysis, the results showed that IL-10 -1082A>G polymorphism was not significantly associated with increased risk of CD under all genetic models genetic models i.e., allele (A vs. G: OR=0.973, 95% CI 0.848-1.116, P=0.693), homozygote (AA vs GG: OR=0.925, 95% CI 0.696-1.230, P=0.594), heterozygote (AG vs GG: OR=0.921, 95% CI 0.618-1.373, P=0.687), dominant (AA+AG vs GG: OR=0.882, 95% CI 0.637-1.221, P=0.449), and recessive (AA vs AG+GG: OR=0.960, 95% CI 0.747-1.234, P=0.752, Figure 2B).
Sensitivity analysis
Sensitivity analyses were performed after sequential removal of each of the included studies and HWE-violating studies to examine the influence of each individual data-set on the pooled ORs. However, the pooled ORs and 95% CIs were not significantly altered when any part of the study was omitted, which indicated that any single study had little impact on the overall ORs. Moreover, we included also those HWE-violating studies for IL-6 -174G>C polymorphism in the current meta-analysis. However, after those studies were excluded, the IL-6 -174G>C polymorphism association with CD risk was not adjusted (data not shown).
Publication bias
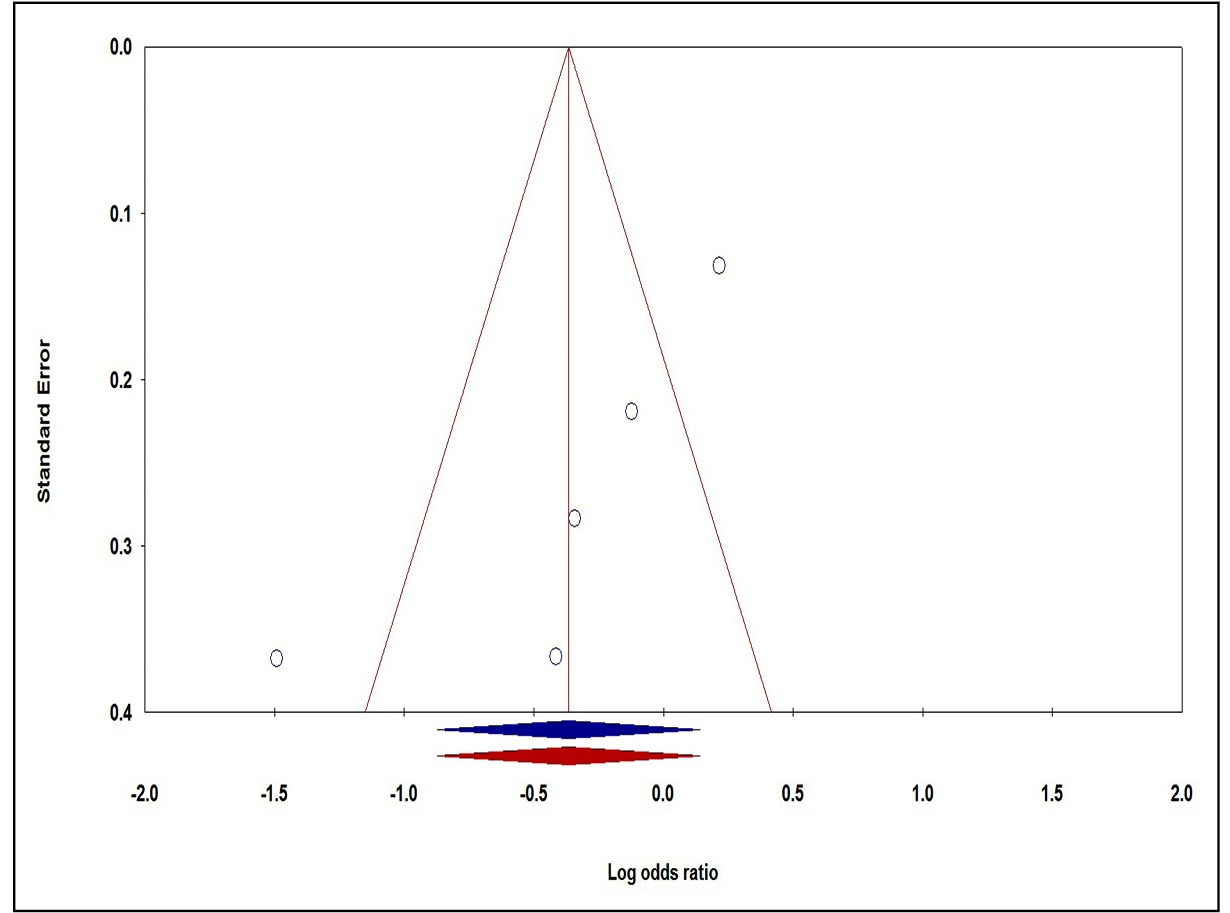
We used visual inspection of funnel plots and Egger’s test to evaluate the publication bias in the current meta-analysis. The shape of funnel plots showed no obvious asymmetry and the result of Egger’s test did not show statistical evidence for bias. The Egger’s test also showed that All the P values were more than 0.05 for IL-10 -1082A>G (Table 2). Thus, no evident publication bias was found in present study, and the result of the study is reliable and credible. However, publication bias in the included studies for IL-6 -174G>C polymorphism showed evidence of an asymmetry under dominant model and supported by Egger’s test (P Beggs=0.086 and P Eggers=0.046, Table 2). Thus, we utilized the Duval and Tweedie non-parametric ‘‘trim and fill’’ method to testing and adjusting the publication bias for IL-6 -174G>C polymorphism. However, the results did not adjust, indicating that our results were statistically robust and reliable (Figure 3).
Funnel plot for publication bias in the meta-analysis of IL-6 -174G>C polymorphism and CD risk under the dominant model (AA+AG vs GG). “Blue” without and “red” with trim and fill method.
DISCUSSION
CD is a long term and complex immune-mediated enteropathy with a very strong genetic component. Multiple genetic findings over the last decade have added to the already known HLA-DQ2 and HLA-DQ8 haplotypes influence numerous loci associated to risk of CD2323. Dieli-Crimi R, Cénit MC, Núñez C. The genetics of celiac disease: A comprehensive review of clinical implications. J Autoimmun. 2015;64:26-41.. In the present study, we performed a comprehensive meta-analysis to evaluate the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms to susceptibility CD based on all available studies. The results demonstrated that the IL-6 -174G>C and IL-10 -1082A>G polymorphisms not significantly associated with risk of CD in overall population. Most of the studies included in this meta-analysis were recruited from Caucasian populations. So, we did not analyze the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD risk by ethnicity, and future studies should address this point.
As a pleiotropic cytokine, IL-6 can regulate many cell functions, including immune defensive mechanism, cell proliferation, and differentiation, and also the production of haemocytes. Previous epidemiological studies have shown an association between IL-6 -174G>C polymorphism and CD risk, but the results are controversial and inconclusive. For example, de Albuquerque et al., in a case-control of 192 CD cases and 96 controls from northeast Italy evaluated the association IL-6 -174G>C polymorphism with CD. Their results showed a statistically significant association between IL-6 -174 G>C polymorphism and CD1818. de Albuquerque Maranhão RM, Martins Esteves FA, Crovella S, Segat L, Eleutério Souza PR. Tumor necrosis factor-α and interleukin-6 gene polymorphism association with susceptibility to celiac disease in Italian patients. Genet Mol Res. 2015;14:16343-52.. Dema et al., have reported that the IL-6 -174G>C polymorphism was significantly associated with increased risk of CD susceptibility in Spanish girls1717. Dema B, Martínez A, Fernández-Arquero M, Maluenda C, Polanco I, Figueredo MA, et al. The IL6-174G/C polymorphism is associated with celiac disease susceptibility in girls. Hum Immunol. 2009;70:191-4.. However, Barartabar et al., not found a significant association between IL-6 -174G>C polymorphism and increased risk of CD in an Iranian population1919. Barartabar Z, Nikzamir A, Sirati-Sabet M, Aghamohammadi E, Chaleshi V, Rostami Nejad M, et al. The relationship between 174 G/C and -572 G/C of IL-6 gene polymorphisms and susceptibility of celiac disease in the Iranian population. Gastroenterol Rev . 2018;13:293-8.. This inconsistency could result from relatively small sample size, from genetic ethnical heterogeneity or from unknown local environmental factors. In current meta-analysis, we pooled all five eligible case-control studies addressing the association of the IL-6 -174 G>C polymorphism with CD, and the results revealed a negative association between this polymorphism and risk of CD.
There has been little evidence to suggest association between the IL-10 -1082A>G polymorphism and CD risk, and their results are inconclusive. Hahn-Zoric et al., have evaluated the association of the IL-10 -1082A>G and TNF-α -308 G>A polymorphisms with CD risk in a Swedish population. Their results showed a possible involvement of IL-10 -1082A>G polymorphism in the development of CD2020. Hahn-Zoric M, Hytönen AM, Hanson LA, Nilsson LA, Padyukov L. Association of -1087 IL10 and -308 TNFA gene polymorphisms with serological markers of coeliac disease. J Clin Immunol. 2003;23:291-6.. Zupin et al., in a study evaluated the association of IL-10 -1082A>G, -819T>C and -592A>C polymorphisms with CD risk in 565 cases and 576 controls from north-eastern Italy2222. Zupin L, Polesello V, Catamo E, Crovella S, Segat L. Interleukin-10 gene promoter polymorphisms in celiac patients from north-eastern Italy. Hum Immunol . 2014;75:656-61.. Their results showed that the IL-10 polymorphisms were not significantly associated with increased risk CD in the population. Lio et al., not found differences in genotype frequencies of IL-10 -1082A>G polymorphism between cases with CD and controls in an Italian population2121. Lio D, Scola L, Forte GI, Accomando S, Giacalone A, Crivello A, et al. TNFα, IFNγ and IL-10 gene polymorphisms in a sample of Sicilian patients with coeliac disease. Dig Liver Dis. 2005;37:756-60.. To statistically discuss this association, we designed the present meta-analysis based on all previously published studies. After data syntheses, we found no significant association between IL-10 -1082A>G polymorphism and the susceptibility to CD in overall population. For the low penetrance CD susceptibility gene effects from SNP, these important genetic and environmental factors should be adequately considered.
To the best of our knowledge, our study is the first meta-analysis focused on the association of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD risk. In addition, the methodological issues for meta-analysis, such as, heterogeneity, publication bias, and sensitivity analysis for stability of results were all carefully investigated. Despite these advantages, some limitations in current study should be acknowledged when interpreting our results. First, this meta-analysis was limited by the small sample size, which needs further investigations. Second, in this meta-analysis study, all case-control studies were recruited from Caucasians and thus, the results may be applicable to the Caucasian populations only. Further studies in others ethnic groups are required to give more comprehensive understanding the exact role of these polymorphisms on CD risk. Third, only published studies were recruited in this meta-analysis, publication bias might have inevitably occurred. Fourth, only articles that were published in English, Persian and Chinese, and studies with available full-text articles were included in this meta-analysis; therefor, some eligible studies that have not been unpublished or were reported in other languages were missed, which may influence the pooled results. Finally, CD is a multifactorial disease that results from complex interactions between genetic and environmental factors. Due to the lack of uniform background data for recruited studies, our results were based on unadjusted estimates, data were not stratified by other factors such as age, gender, lifestyle factors, environment factors, and other risk factors of CD risk. With more data becoming available, it might be evaluate the association with more reliable results.
In summary, our pooled data indicated that the IL-6 -174G>C and IL-10 -1082A>G polymorphisms are not an important susceptibility genetic factor for CD development. However, to reach a definitive conclusion, further well-designed studies with larger sample size, detailed data and in others ethnic groups are required to further clarify the exact role of IL-6 -174G>C and IL-10 -1082A>G polymorphisms with CD risk. Moreover, gene-gene and gene-environment interactions studies should also be considered in future studies.
REFERENCES
-
1Rossi E, Basso D, Zambon C-F, Navaglia F, Greco E, Pelloso M, et al. TNFA Haplotype Genetic Testing Improves HLA in Estimating the Risk of Celiac Disease in Children. Goldberg AC, editor. PLoS One. 2015;10:e0123244.
-
2Barartabar Z, Nikzamir A, Sirati-Sabet M, Aghamohammadi E, Chaleshi V, Rostami Nejad M, et al. The relationship between 174 G/C and -572 G/C of IL-6 gene polymorphisms and susceptibility of celiac disease in the Iranian population. Gastroenterol Rev. 2018;13:293-8.
-
3Aflatoonian M, Moghimi M, Akbarian-Bafghi MJ, Morovati-Sharifabad M, Jarahzadeh MH, Neamatzadeh H. Association of TNF- α-308G>A polymorphism with susceptibility to celiac disease: a systematic review and meta-analysis. Arq Gastroenterol. 2019;56:88-94.
-
4Dubé C, Rostom A, Sy R, Cranney A, Saloojee N, Garritty C, et al. The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology. 2005;128(4 Suppl 1):S57-67.
-
5Cheng J, Brar PS, Lee AR, Green PHR. Body Mass Index in Celiac Disease. J Clin Gastroenterol. 2010;44:267-71.
-
6Capilla A, Donat E, Planelles D, Espinós C, Ribes-Koninckx C, Palau F. Genetic analyses of celiac disease in a Spanish population confirm association with CELIAC3 but not with CELIAC4. Tissue Antigens. 2007;70:324-9.
-
7Khan S, Mandal RK, Jawed A, Dar SA, Wahid M, Panda AK, et al. TNF-α -308 G > A (rs1800629) Polymorphism is Associated with Celiac Disease: A Meta-analysis of 11 Case-Control Studies. Sci Rep. 2016;6:32677.
-
8Akobeng AK, Ramanan A V, Buchan I, Heller RF. Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies. Arch Dis Child. 2005;91:39-43.
-
9Broce I, Karch CM, Wen N, Fan CC, Wang Y, Hong Tan C, et al. Immune-related genetic enrichment in frontotemporal dementia: An analysis of genome-wide association studies. Brayne C, editor. PLOS Med. 2018;15:e1002487.
-
10Ren H, Zhang Y, Yao Y, Guo T, Wang H, Mei H, et al. Association between the interleukin-6 genetic polymorphism 174 G/C and thrombosis disorder risk. Medicine (Baltimore). 2016;95:e4030.
-
11Namazi A, Forat-Yazdi M, Jafari M, Farahnak S, Nasiri R, Foroughi E, et al. Association of Interleukin-10 -1082 a/G (Rs1800896) Polymorphism With Susceptibility To Gastric Cancer: Meta-Analysis of 6,101 Cases and 8,557 Controls. Arq Gastroenterol . 2018;55:33-40.
-
12Sheikhpour E, Noorbakhsh P, Foroughi E, Farahnak S, Nasiri R, Neamatzadeh H. A survey on the role of interleukin-10 in breast cancer: A narrative. Reports Biochem Mol Biol. 2017;7(1).
-
13Hirano T. Interleukin 6 in autoimmune and inflammatory diseases: a personal memoir. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:717-30.
-
14Beebe AM, Cua DJ, de Waal Malefyt R. The role of interleukin-10 in autoimmune disease: systemic lupus erythematosus (SLE) and multiple sclerosis (MS). Cytokine Growth Factor Rev. 2002;13:403-12.
-
15Garrote JA, Arranz E, Tellería JJ, Castro J, Calvo C, Blanco-Quirós A. TNF alpha and LT alpha gene polymorphisms as additional markers of celiac disease susceptibility in a DQ2-positive population. Immunogenetics. 2002;54:551-5.
-
16Barisani D, Ceroni S, Meneveri R, Cesana BM, Bardella MT. IL-10 polymorphisms are associated with early-onset celiac disease and severe mucosal damage in patients of Caucasian origin. Genet Med. 2006;8:169-74.
-
17Dema B, Martínez A, Fernández-Arquero M, Maluenda C, Polanco I, Figueredo MA, et al. The IL6-174G/C polymorphism is associated with celiac disease susceptibility in girls. Hum Immunol. 2009;70:191-4.
-
18de Albuquerque Maranhão RM, Martins Esteves FA, Crovella S, Segat L, Eleutério Souza PR. Tumor necrosis factor-α and interleukin-6 gene polymorphism association with susceptibility to celiac disease in Italian patients. Genet Mol Res. 2015;14:16343-52.
-
19Barartabar Z, Nikzamir A, Sirati-Sabet M, Aghamohammadi E, Chaleshi V, Rostami Nejad M, et al. The relationship between 174 G/C and -572 G/C of IL-6 gene polymorphisms and susceptibility of celiac disease in the Iranian population. Gastroenterol Rev . 2018;13:293-8.
-
20Hahn-Zoric M, Hytönen AM, Hanson LA, Nilsson LA, Padyukov L. Association of -1087 IL10 and -308 TNFA gene polymorphisms with serological markers of coeliac disease. J Clin Immunol. 2003;23:291-6.
-
21Lio D, Scola L, Forte GI, Accomando S, Giacalone A, Crivello A, et al. TNFα, IFNγ and IL-10 gene polymorphisms in a sample of Sicilian patients with coeliac disease. Dig Liver Dis. 2005;37:756-60.
-
22Zupin L, Polesello V, Catamo E, Crovella S, Segat L. Interleukin-10 gene promoter polymorphisms in celiac patients from north-eastern Italy. Hum Immunol . 2014;75:656-61.
-
23Dieli-Crimi R, Cénit MC, Núñez C. The genetics of celiac disease: A comprehensive review of clinical implications. J Autoimmun. 2015;64:26-41.
-
Disclosure of funding: no funding received
Publication Dates
-
Publication in this collection
30 Sept 2019 -
Date of issue
Jul-Sep 2019
History
-
Received
29 Apr 2019 -
Accepted
15 July 2019