Abstract
Forty-nine isolates of Trichoderma from the Brazilian Midwest were evaluated for their antagonistic activity in vitro against Sclerotinia sclerotiorum (causal agent of white mold), which were then identified based on their nuclear ribosomal ITS sequences. Paired culture tests showed that all isolates exhibited some antagonism, with a maximum of 77% mycelial inhibition and complete inhibition of sclerotia production. Two isolates were found to be the most promising biocontrol agents, considering both antagonistic parameters (CEN1253 - T. koningiopsis and CEN1265 - T. brevicompactum). Five different species were identified: T. harzianum (23), T. spirale (9), T. koningiopsis (8), T. brevicompactum (7) and T. asperellum (2). These isolates are stored in the Embrapa Fungi Collection for Biological Control and the information obtained in the experiments will be incorporated into the database of biological assets within the genetic resources information system (Allele) and be made available for further studies.
Keywords
Microbial genetic resources; molecular identification; biocontrol; white mold
Resumo
Quarenta e nove isolados de Trichoderma obtidos no centro-oeste do Brasil foram avaliados quanto a sua atividade antagônica in vitro contra Sclerotinia sclerotiorum (agente causal do mofo branco) e identificados com base nas sequências ITS do DNA ribossômico nuclear. Os testes de cultivo pareado mostram que todos os isolados exibiram algum antagonismo, com um máximo de 77% de inibiação micelial e inibição total da produção de escleródios. Dois isolados se destacaram como os mais promissores, considerando ambos os parâmetros avaliados (CEN1253 - T. koningiopsis e CEN1265 - T. brevicompactum). Cinco espécies diferentes foram identificadas: T. harzianum (23), T. spirale (9), T. koningiopsis (8), T. brevicompactum (7) and T. asperellum (2). Estes isolados estão armazenados na Coleção de Fungos para Controle Biológico da Embrapa e as informações obtidas nos experimentos serão incorporadas na base de dados de ativos biológicos, no sistema de informações de recursos genéticos, e disponibilizados para estudos futuros.
Palavras-chave
Recursos genéticos microbianos; identificação molecular; biocontrole; mofo branco
Introduction
Sclerotinia sclerotiorum Lib. de Bary (Ss) is the causal agent of white mold, a disease that affects many hosts such as legumes, sunflowers, canola, most vegetables, and others. This pathogen is widespread in Brazil and the fungus can cause serious losses, mainly due to its contamination or infestation of seeds (Heffer Link & Johnson 2007HEFFER LINK, V. & JOHNSON, K.B. 2007. White Mold. The Plant Health Instructor, http://dx.doi.org/10.1094/PHI-I-2007-0809-01
http://dx.doi.org/10.1094/PHI-I-2007-080...
). Furthermore, the pathogen survives for long periods using specialized resistant structures, called sclerotia (Bianchini et al. 2005BIANCHINI, A., MARINGONI, A.C. & CARNEIRO, S.M.T.G. 2005. Doenças do feijoeiro (Kimati et al., Eds.). In Manual de Fitopatologia. Vol. 2. São Paulo: Agronômica Ceres. p.333-360.). For soil pathogens, chemical control rarely leads to satisfactory results. Furthermore, chemical pesticides pose health risks, lead to the reduction of beneficial soil microflora and increase production costs (Fahmi et al. 2012FAHMI, A.I., AL-TAHHI, A.D. & HASSAN, M.M. 2012. Protoplast fusion enhances antagonistic activity in Trichoderma sp Nat & Science 10(5):100-106.). In this context, biological control of plant diseases using antagonists has already shown good results in Brazil, as in other countries with more tradition in this practice, such as USA, Canada, Australia and France (Campanhola & Bettiol 2003CAMPANHOLA, C., & BETTIOL, W. 2003. Métodos alternativos de controle fitossanitário. Brasília: Embrapa Informação Tecnológica. 279p.). Particularly in the areas of organic production, there is high demand for biological control agents, because they have a low impact on the environment, leave no toxic residues in food and are fully compatible with other alternative control measures. However, one of the keys to success in the use of biofungicides is that they must be developed from strains with high activity against the pathogens in question and be adapted to the environmental conditions under which they will operate.
It is necessary to maintain a continuous flow of collection, isolation, evaluation and characterization of biocontrol agents to improve and maintain their efficiency (Lopes et al. 2013LOPES, R.B, BRITO, M.A.V., MELLO, S.C.M. & SAGGIN, O.J. 2013. Coleções microbianas na Embrapa: conservação e agregação de valor à biodiversidade, In: Simpósio Microrganismos em Agroenergia: da Prospecção aos Bioprocessos. Almeida JRM (Ed.). Brasília, DF: Embrapa Agroenergia (Documentos/Embrapa Agroenergia, ISSN 2177- 4439). p.15-22.). Species of Trichoderma Pers. are widely used in the biological control of plant pathogens, but commercial formulations have only become available recently. The lack of properly registered formulations has been a serious obstacle in the use of bioproducts in Brazil (Machado et al. 2012MACHADO, D.F.M., PARZIANELLO, R., SILVA, A.C.F. & ANTONIOLLI, Z.I. 2012. Trichoderma no Brasil: O Fungo e o Bioagente. Ciências Agrárias 35(1):274-288.). According to the Brazilian Ministry of Agriculture Livestock and Food Supply (MAPA) database (Agrofit 2016AGROFIT. Busca por produtos formulados.http://agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons(last acess: 02/02/2016)
http://agrofit.agricultura.gov.br/agrofi...
), there are five commercial products recommended for Ss control, formulated with either T. asperellum and T. harzianum. In order to select isolates for new formulations, some studies have reported the effectiveness of different species of Trichoderma in control of Ss, showing both inhibition of mycelial growth (Amin et al. 2010AMIN, F., RAZDAN, V.K., MOHIDDIN, F.A., BHAT, K.A. & BANDAY, S. 2010. Potential of Trichoderma species as biocontrol agents of soil borne fungal propagules. J Phytol 2(10):38-41., Samuel et al. 2010SAMUELS, G.J., ISMAIEL, A., BOM, M.C., DE RESPINIS, S. & PETRINI, O. 2010. Trichoderma asperellum sensu lato consists of two crypitc species. Mycologia 104(4):944-966, http://dx.doi.org/10.3852/09-243
http://dx.doi.org/10.3852/09-243...
, Matroudi et al. 2009MATROUDI, I.S., ZAMANI, M.R. & MOTALLEBI, M. 2009. Antagonistic effects of three species of Trichoderma sp. on Sclerotinia sclerotiorum, the causal agent of canola stem rot. Egypt J Biol 11:37-44, http://dx.doi.org/10.4314/ejb.v11i1.56560
http://dx.doi.org/10.4314/ejb.v11i1.5656...
) and the parasitism of sclerotia (Abdullah et al. 2008ABDULLAH, M.T., ALI, N.Y., & SULEMAN P. 2008. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary with Trichoderma harzianum and Bacillus amyloliquefaciens. Crop Prot 27(10):1354-1359, http://dx.doi.org/10.1016/j.cropro.2008.05.007
http://dx.doi.org/10.1016/j.cropro.2008....
).
The current taxonomy of Trichoderma is based mainly on molecular analysis (Druzhinina et al. 2010DRUZHININA, I.S, KUBICEK, C.P., KOMOŃ-ZELAZOWSKA, M., MULAW, T.B & BISSETT, J. 2010. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol 10:1-14, http://dx.doi.org/10.1186/1471-2148-10-94
http://dx.doi.org/10.1186/1471-2148-10-9...
, Kullning et al. 2000), since morphological identification of the anamorph forms are unreliable (Druzhinina et al. 2006DRUZHININA, I.S, KOPCHINSKIY, A. & KUBICEK, C.P. 2006. The first 100 Trichoderma species characterized by molecular data. Mycoscience 47:55-64, http://dx.doi.org/10.1007/s10267-006-0279-7
http://dx.doi.org/10.1007/s10267-006-027...
). With the development of molecular phylogeny, the genus Trichoderma has been grouped into taxonomic sections (Bissett 1984BISSETT, J. 1984. A revision of the genus Trichoderma. I. Sect. Longibrachiatum sect. nov. Can J Bot. 62:924-931.); based on phylogenetic clades (Druzhinina et al. 2010DRUZHININA, I.S, KUBICEK, C.P., KOMOŃ-ZELAZOWSKA, M., MULAW, T.B & BISSETT, J. 2010. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol 10:1-14, http://dx.doi.org/10.1186/1471-2148-10-94
http://dx.doi.org/10.1186/1471-2148-10-9...
). As an extension of this work, Druzhinina and colleagues (2005) developed a DNA barcode for molecular identification of Trichoderma species, called TrichOKEY. Given the importance of biological control within the sustainable management of agriculture and environmental protection, the current investigation was undertaken to prospect for Trichoderma isolates with potential for inhibition of Ss in soil of the Federal District, Brazil and to identify them using nrRNA gene ITS sequences.
Material and Methods
The samples were obtained from soils from organic farming systems and native vegetation, from Rural Center Rajadinha (Planaltina, Federal District). The region is part of the Brazilian central highlands, with an altitude of around 954 m. The main soil type that predominates in the region is red latosol. The collected soils were extracted from fractions of rhizoplane, rhizosphere and roots, up to 5 cm from the following plants: maize (Zea mays L.); eggplant (Solanum melongena L.); okra (Abelmoschus esculentus L. Moench); tomato (Solanum lycopersicum L.); squash (Cucurbita sp.); cassava (Manihot esculenta Crantz); cabbage (Brassica oleracea var. capitata L.); ornamental fern (Pleopeltis sp.), kale (Brassica oleracea L. var. acephala DC.); Cestrum sp. and Miconia sp. (Table 2). Pure cultures of the antagonist, originating from monosporic cultures, were tested against Ss strain CEN1157 in paired culture experiments, according to Dennis & Webster (1971)DENNIS, C. & WEBSTER, J. 1971. Antagonistic properties of species-groups of Trichoderma. III. Hyphal interactionsT Brit Mycol Soc 57(3):363-369, http://dx.doi.org/10.1016/S0007-1536(71)80077-3
http://dx.doi.org/10.1016/S0007-1536(71)...
. The plates were incubated aerobically for seven days at 25 °C, in a photoperiod of 12 hours.
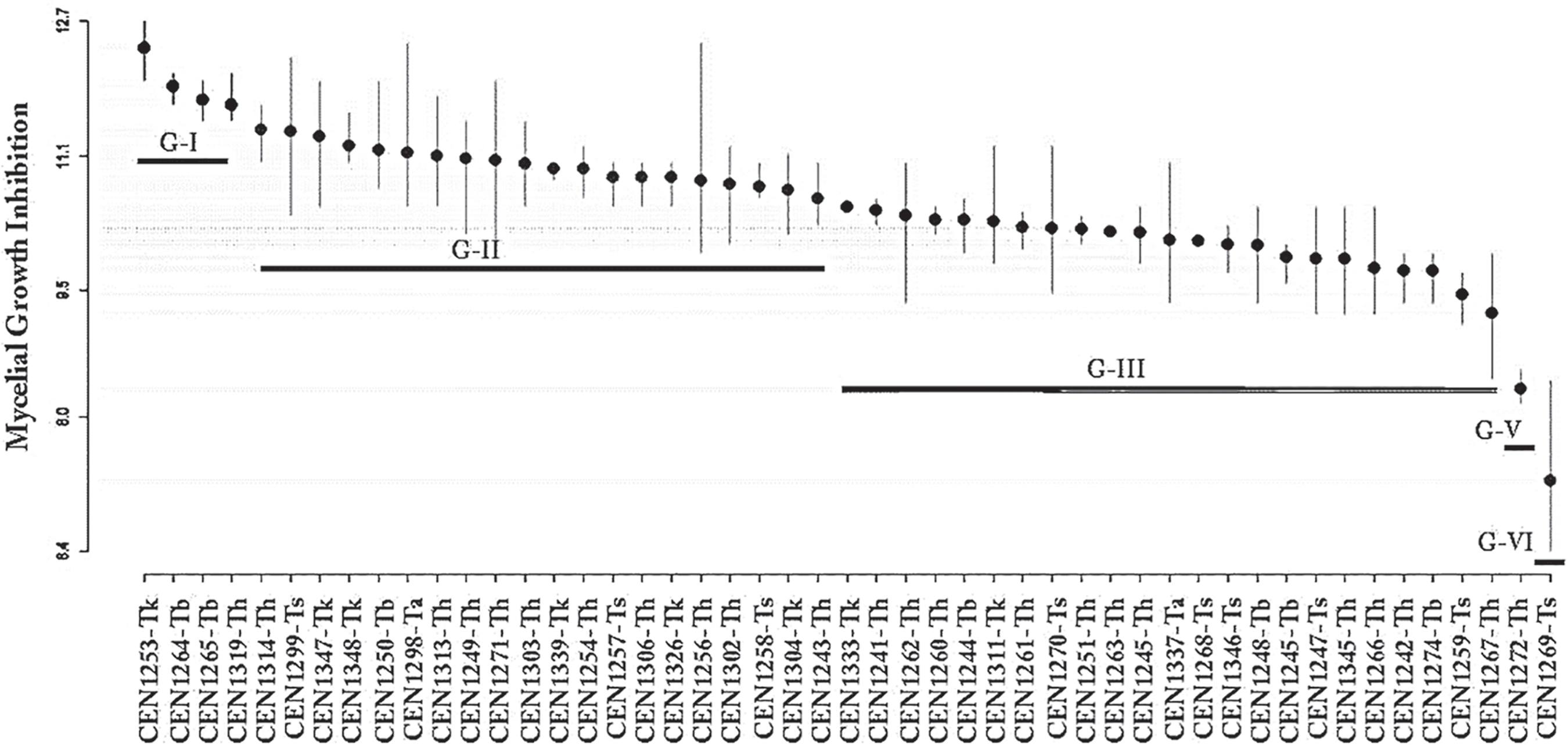
List of the Trichoderma isolates, results of the in vitro mycelial growth inhibition test (MGI) percentage and number of sclerotia (NS) produced by Sclerotinia sclerotiorum.
The confrontation tests were performed in triplicate and the control was a disc of culture medium with Ss in the center of plate. Radial mycelial growth of the pathogen (cm) was measured with the aid of a millimeter ruler and used for calculating inhibition of mycelial growth rate (Menten et al. 1976MENTEN, J.O.M., MINUSSI, C.C., CASTRO, C. & KIMATI, H. 1976. Efeito de alguns fungicidas no crescimento micelial de Macrophomina phaseolina (Tass.) Goid. "in vitro". Fitopatol Bras 1(2):57-66, http://dx.doi.org/10.1590/S1983-40632013000400014
http://dx.doi.org/10.1590/S1983-40632013...
) using the equation: MGI = [(Dtest-Dtrat/Dtrat)*100]; where Dtest is the diameter of the radial mycelial growth of Ss in the control treatment without Trichoderma), Dtrat, is the diameter of the radial mycelial growth of Ss in treatment with Trichoderma. The number of sclerotia produced by Ss was also counted in the confrontation cultures zones.
Comparison of means was performed using the Scott-Knott clustering test. The model residues were subjected to Box-Cox transformation when they had normal variance. Box-Cox consisted of transforming the data according to expression: Y'= (Yλ-1)/λ, where Y is the response variable under investigation and λ is a parameter to be estimated. Analyses were performed in the statistical program R (http://wwwr-project.org/). The significance level for all analyses was 5%.
Genomic DNA was extracted using a phenol-chloroform method (Raeder & Broda 1985RAEDER, U. & BRODA, P. 1985. Rapid preparation of DNA from filamentous fungiLett Appl Microbiol 1(1):17-20, http://dx.doi.org/10.1111/j.1472-765X.1985.tb01479.x
http://dx.doi.org/10.1111/j.1472-765X.19...
) from Trichoderma isolates grown in potato dextrose broth in a shaking incubator at 150 rpm for five days. The gDNA was quantified by comparison to a high molecular weight DNA Mass Ladder (Invitrogen). The nuclear ribossomal ITS locus (rDNA; ITS1-5.8S-ITS2) was amplified by PCR using the primers ITS1 5'-TTCCGTAGGTGAACCTGCGG-3' and ITS4 5'-TCCTCCGCTTATTGATATGC-3' (White et al. 1990WHITE, T.J., BRUNS, T., LEE, S. & TAYLOR, J.W. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, p.315-322, http://dx.doi.org/10.1016/B978-0-12-372180-8.50042-1
http://dx.doi.org/10.1016/B978-0-12-3721...
). The PCR reaction volume was 25 µL and contained: 5 ng template DNA; 1 X PCR buffer; 0.2 mM dNTPs; 1.5 mM MgCl2; 0.4 µM each primer; 1 U Taq DNA polymerase. PCR cycling consisted of an initial 96 °C for 5 min, then 30 cycles 94 °C for 45 s, 60 °C for 45 s and 72 °C for 1 min with a final extension at 72 °C for 10 min. PCR products were visualized using 1% agarose gels and the products subjected to Sanger chain termination sequencing (both strands) using the amplification primers (Macrogen Inc.; Seoul, South Korea).
Contigs were assembled using DNA BASER (http://www.dnabaser.com/index.html). BLAST (Basic Local Alignment Search Tool) searches of the NCBI (National Center for Biotechnology Information) Genbank nucleotide database and TrichOKEY 2.0 (http://www.isth.info/) were used to verify the ITS sequences. Experimentally derived sequences and related reference ITS sequences obtained from Genbank were aligned using MAFFT with its G-INSI option (Katoh & Standley 2013KATOH, K. & STANDLEY, D.M. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30(4):772-780, http://dx.doi.org/10.1631/jzus.B0860015
http://dx.doi.org/10.1631/jzus.B0860015...
). A phylogenetic hypothesis was calculated using MrBayes (v3.2.5; Ronquist & Huelsenbeck 2003RONQUIST, F. & HUELSENBECK, J.P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:72-4, http://dx.doi.org/10.1093/bioinformatics/btg180
http://dx.doi.org/10.1093/bioinformatics...
). The Bayesian analysis was conducted using default program settings except that reversible-jump MCMC (Huelsenbeck et al. 2004HUELSENBECK, J.P., LARGET, B. & ALFARO, M.E. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol 21(6):1123-1133, http://dx.doi.org/10.1093/molbev/msh123
http://dx.doi.org/10.1093/molbev/msh123...
) was used to optimize base substitution model parameters within the general time reversible (GTR) framework. MCMC sampling was run for 2 million generations, which allowed the standard deviation of split frequencies to fall to 0.006853, strongly indicating stable convergence. The analysis was repeated to confirm the result obtained. The first 25% of trees were discarded as burn-in prior to calculation of the 50% majority rule consensus phylogram. The ITS sequence from T. longibrachiatum (Table 1) was used as outgroup for tree rooting.
Results
In the paired culture experiments, we observed inhibition of mycelial growth of Ss ranging between 28, and 77% (Table 2).
In the statistical analysis of MGI, we observed the formation of five distinct groups (horizontal bars; Figure 1) at the 5% significance level in the Scott-Knott test, as shown (Figure 1). The data were transformed (Box-Cox, λ = 0.42) and the model was able to explain 91.69% of the total variability in the data (SQResidues= 33.14, SQTreat= 109.98, R2=69%, F48,98= 6.775, p-value= 7.8e-16).
Average inhibition of mycelial growth of Sclerotinia sclerotiorum by Trichoderma spp., according to the Scott-Knott test.
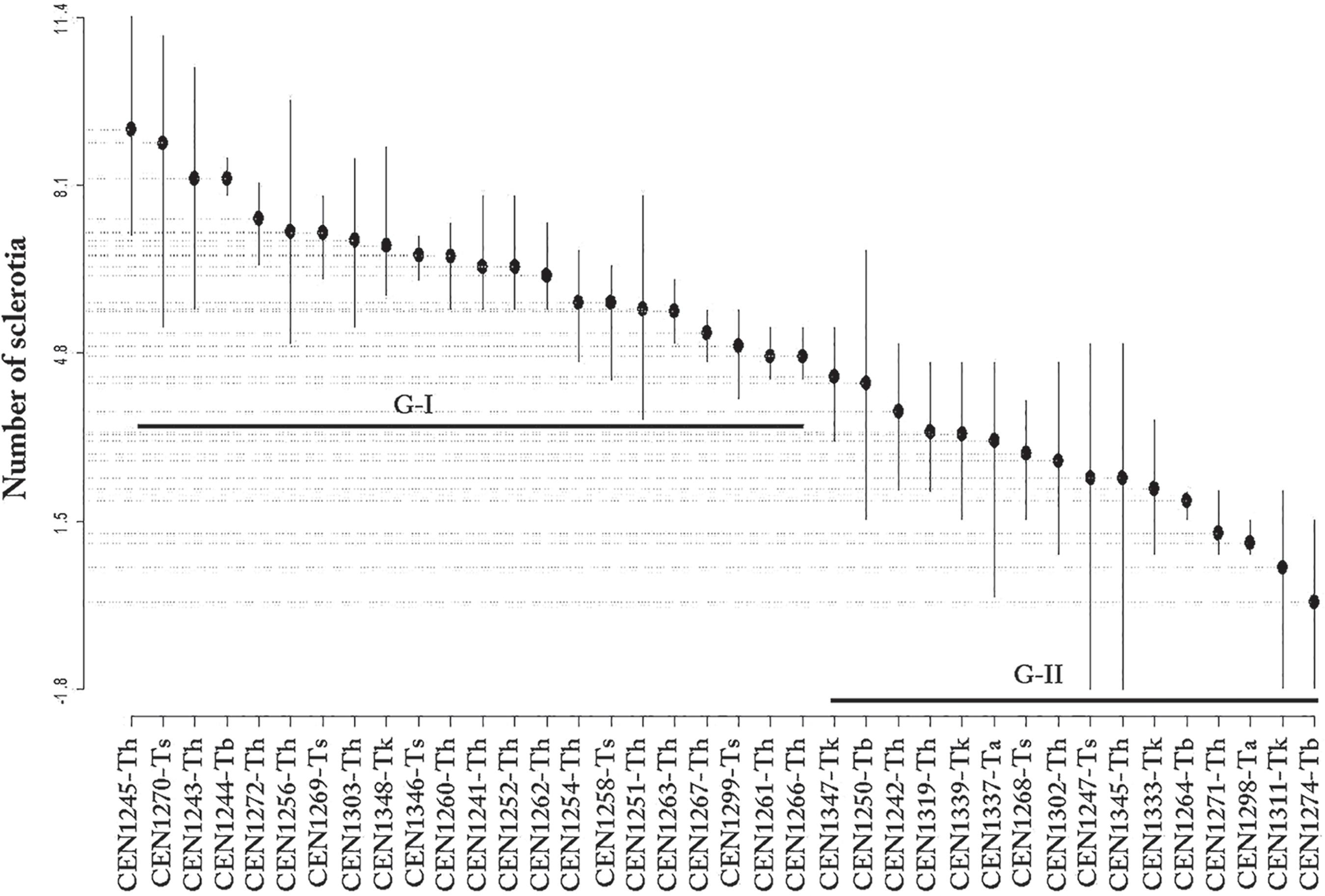
Production of Ss sclerotia was also reduced in the confrontation cultures by the action of Trichoderma isolates (Figure 2). At the 5% level of significance, the ScottKnott test showed that the isolates formed two separate groups. The data were transformed (Box-Cox, λ = 0.55) and the model was able to explain 66% of the total variability in the data (SQResidues= 222.6, SQTreat= 660.3, R2=66%, F37,76= 6.092, p-value < 1.61e-11). The isolates CEN1253, CEN1314, CEN1313, CEN1257 and CEN1304 all caused complete inhibition of sclerotia production (Table 2). The first group of 25 isolates had less inhibition of sclerotia production, ranging between 27 and 10 sclerotia, while in the second group, between eight and two sclerotia were produced.
Mean production of Ss sclerotia in the presence of Trichoderma, according to the Scott-Knott test.
In the phylogenetic analysis of ITS sequences, rooted with T. longibrachiatum, we observed five major clades (Figure 3). The first 23 isolates grouped with the T. harzianum reference with a posterior probability of one, although some variability within the clade was present. The second clade contained nine isolates clustering with T. spirale, again with a posterior probability of one. The third group comprised into two related clades, one containing eight isolates and T. koningiopsis, and the other two isolates clustering with T. asperellum. Finally, the fifth clade contained seven isolates and T. brevicompactum.
Discussion
In the present work, isolates with greater inhibitory potential were found to belong to the species T. koningiopsis, T. brevicompactum and T. harzianum. Experiments carried out by Amin et al. (2010)AMIN, F., RAZDAN, V.K., MOHIDDIN, F.A., BHAT, K.A. & BANDAY, S. 2010. Potential of Trichoderma species as biocontrol agents of soil borne fungal propagules. J Phytol 2(10):38-41. showed that T. viride was most inhibitory to S. sclerotiorum (66%), while two isolates of T. harzinaum exhibited MGI values of 56.57% and 38.12% respectively. Matroudi et al. (2009)MATROUDI, I.S., ZAMANI, M.R. & MOTALLEBI, M. 2009. Antagonistic effects of three species of Trichoderma sp. on Sclerotinia sclerotiorum, the causal agent of canola stem rot. Egypt J Biol 11:37-44, http://dx.doi.org/10.4314/ejb.v11i1.56560
http://dx.doi.org/10.4314/ejb.v11i1.5656...
used an Ss isolate from canola, to evaluate antagonistic Trichoderma spp., reporting variation between isolates. However, T. atroviride was the most efficient species and reduced growth between 85-93% compared to T. harzianum that presented an MGI of 70-80%. Likewise, there was no correlation between the source niche of the antagonist and its inhibitory potential in vitro. Here, the most efficient isolates were obtained from both the rhizosphere and rhizoplane as well from soil samples.
The ability to reduce the production of sclerotia is a valuable quality for the selection of Trichoderma isolates for biocontrol in view of the importance of the resistant structure on the survival of the pathogen in soil (Bianchini et al. 2005BIANCHINI, A., MARINGONI, A.C. & CARNEIRO, S.M.T.G. 2005. Doenças do feijoeiro (Kimati et al., Eds.). In Manual de Fitopatologia. Vol. 2. São Paulo: Agronômica Ceres. p.333-360.). Some isolates from this study (T. koningiopsis, T. harzinaum and T. spirale) completely inhibited the production of Ss sclerotia. In the same sense, Abdullah et al. (2008)ABDULLAH, M.T., ALI, N.Y., & SULEMAN P. 2008. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary with Trichoderma harzianum and Bacillus amyloliquefaciens. Crop Prot 27(10):1354-1359, http://dx.doi.org/10.1016/j.cropro.2008.05.007
http://dx.doi.org/10.1016/j.cropro.2008....
observed inhibition of the formation of Ss sclerotia by T. harzianum, that decreased from 31.66 (treatment control) to 12.07 and 18.12 sclerotia. On the other hand, the most inhibitory isolate from the work reported by Amin et al. (2010)AMIN, F., RAZDAN, V.K., MOHIDDIN, F.A., BHAT, K.A. & BANDAY, S. 2010. Potential of Trichoderma species as biocontrol agents of soil borne fungal propagules. J Phytol 2(10):38-41. was T. viride.
Following the classification of Druzhinina et al. (2010)DRUZHININA, I.S, KUBICEK, C.P., KOMOŃ-ZELAZOWSKA, M., MULAW, T.B & BISSETT, J. 2010. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol 10:1-14, http://dx.doi.org/10.1186/1471-2148-10-94
http://dx.doi.org/10.1186/1471-2148-10-9...
, we observed some ITS sequence variation among the isolates clustering with T. harzianum. The formation of subgroups within T. harzianum is recognized, since it is regarded as a species complex (Kullnig et al. 2000KULLNIG, C.M., SZAKACS, G. & KUBICEK, C.P. 2000. Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol Res 104:1117-1125, http://dx.doi.org/10.1017/S0953756200002604
http://dx.doi.org/10.1017/S0953756200002...
). The second clade included T. spirale and these first two species have phylogenetic affinity (Druzhinina & Kubicek 2005DRUZHININA, I.S & KUBICEK, C.P. 2005. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? J Zhejiang Univ Sci B 6(2):100-112, http://dx.doi.org/10.1631/jzus.2005.B0100
http://dx.doi.org/10.1631/jzus.2005.B010...
), belonging to Trichoderma Section Pachybasium B. The third group (T. koningiopisis and T. asperellum) with related clades are classified in Section Pachybasium A. The fifth and final clade contained T. brevicompactum are placed in Section Lutea (http://www.isth.info/biodiversity/). Although the ITS sequences showed multiple hits in BLASTn for each query, these were clarified and confirmed by the TrichOKEY program which corroborates the phylogenetic analysis. The advantage of this online tool is that it was developed by taxonomists skilled in Trichoderma/Hypocrea, where the database is restricted to correctly identified sequences (Druzhinina et al. 2005DRUZHININA, I.S, KOPCHINSKIY, A.G., KOMON, M., BISSETT, J., SZAKACS, G. & KUBICEK, C.P. 2005. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol 42(10):813-828, http://dx.doi.org/10.1016/j.fgb.2005.06.007
http://dx.doi.org/10.1016/j.fgb.2005.06....
).
A majority of the isolates were identified as T. harzianum, followed by T. spirale, T. koningiopsis, T. brevicompactum and T. asperellum, respectively. This distribution was expected, since it is known that T. harzianum is a global species, colonizing the most diverse substrates and ecological niches with a broad geographic distribution (Kubicek et al. 2008KUBICEK, C.P., KOMON-ZELAZOWSKA, M. & DRUZHININA, I.S. 2008. Fungal genus Hypocrea/ Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B 9(10):753-763, http://dx.doi.org/10.1631/jzus.B0860015
http://dx.doi.org/10.1631/jzus.B0860015...
).
This study permitted the selection of isolates with good antagonistic potential for S. sclerotiorum and two of them: CEN1253 and CEN1265, identified as T. koningiopsis and T. brevicompactum, respectively, were considered to be promising for both biocontrol parameters (inhibition of mycelial growth and reduction of sclerotia). These fungi are stored in the Embrapa Fungi Collection for Biological Control and the information obtained in the experiments will be incorporated into the database of biological assets and genetic resources system (Allele, http://alelomicro.cenargen.embrapa.br/) for further studies required for the process of development of commercial products and procedures established by the Brazilian Ministry of Agriculture Livestock and Food Supply, until they are available for use by farmers.
Acknowledgements
The authors acknowledge the financial support granted by CAPES and FAP-DF.
References
- ABDULLAH, M.T., ALI, N.Y., & SULEMAN P. 2008. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary with Trichoderma harzianum and Bacillus amyloliquefaciens Crop Prot 27(10):1354-1359, http://dx.doi.org/10.1016/j.cropro.2008.05.007
» http://dx.doi.org/10.1016/j.cropro.2008.05.007 - AGROFIT. Busca por produtos formulados.http://agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons(last acess: 02/02/2016)
» http://agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons - AMIN, F., RAZDAN, V.K., MOHIDDIN, F.A., BHAT, K.A. & BANDAY, S. 2010. Potential of Trichoderma species as biocontrol agents of soil borne fungal propagules. J Phytol 2(10):38-41.
- BIANCHINI, A., MARINGONI, A.C. & CARNEIRO, S.M.T.G. 2005. Doenças do feijoeiro (Kimati et al., Eds.). In Manual de Fitopatologia. Vol. 2. São Paulo: Agronômica Ceres. p.333-360.
- BISSETT, J. 1984. A revision of the genus Trichoderma I. Sect. Longibrachiatum sect. nov. Can J Bot. 62:924-931.
- CAMPANHOLA, C., & BETTIOL, W. 2003. Métodos alternativos de controle fitossanitário. Brasília: Embrapa Informação Tecnológica. 279p.
- DEGENKOLB, T., DIECKMANN, R., NIELSEN, K.F., GRAFENHAN, T., THEIS, C., ZAFARI, D., CHAVERRI, P., ISMAIEL, A., BRUCKNER, H., VON DOHREN, H., THRANE, U., PETRINI, O, & SAMUELS, G.J. 2008. The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol Prog 7(3):177-219, http://dx.doi.org/10.1007/s11557-008-0563-3
» http://dx.doi.org/10.1007/s11557-008-0563-3 - DENNIS, C. & WEBSTER, J. 1971. Antagonistic properties of species-groups of Trichoderma III. Hyphal interactionsT Brit Mycol Soc 57(3):363-369, http://dx.doi.org/10.1016/S0007-1536(71)80077-3
» http://dx.doi.org/10.1016/S0007-1536(71)80077-3 - DRUZHININA, I.S., KOMON-ZELAZOWSKA, M., KREDICS, L., HATVANI, L., ANTAL, Z., BELAYNEH, T., & KUBICEK, C.P. 2008. Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum both capable of causing invasive mycoses of humans. Microbiology 154(11):3447-3459, http://dx.doi.org/10.1099/mic.0.2008/021196-0
» http://dx.doi.org/10.1099/mic.0.2008/021196-0 - DRUZHININA, I.S, KOPCHINSKIY, A. & KUBICEK, C.P. 2006. The first 100 Trichoderma species characterized by molecular data. Mycoscience 47:55-64, http://dx.doi.org/10.1007/s10267-006-0279-7
» http://dx.doi.org/10.1007/s10267-006-0279-7 - DRUZHININA, I.S, KOPCHINSKIY, A.G., KOMON, M., BISSETT, J., SZAKACS, G. & KUBICEK, C.P. 2005. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol 42(10):813-828, http://dx.doi.org/10.1016/j.fgb.2005.06.007
» http://dx.doi.org/10.1016/j.fgb.2005.06.007 - DRUZHININA, I.S, KUBICEK, C.P., KOMOŃ-ZELAZOWSKA, M., MULAW, T.B & BISSETT, J. 2010. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol 10:1-14, http://dx.doi.org/10.1186/1471-2148-10-94
» http://dx.doi.org/10.1186/1471-2148-10-94 - DRUZHININA, I.S & KUBICEK, C.P. 2005. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? J Zhejiang Univ Sci B 6(2):100-112, http://dx.doi.org/10.1631/jzus.2005.B0100
» http://dx.doi.org/10.1631/jzus.2005.B0100 - FAHMI, A.I., AL-TAHHI, A.D. & HASSAN, M.M. 2012. Protoplast fusion enhances antagonistic activity in Trichoderma sp Nat & Science 10(5):100-106.
- HEFFER LINK, V. & JOHNSON, K.B. 2007. White Mold. The Plant Health Instructor, http://dx.doi.org/10.1094/PHI-I-2007-0809-01
» http://dx.doi.org/10.1094/PHI-I-2007-0809-01 - HUELSENBECK, J.P., LARGET, B. & ALFARO, M.E. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol 21(6):1123-1133, http://dx.doi.org/10.1093/molbev/msh123
» http://dx.doi.org/10.1093/molbev/msh123 - KATOH, K. & STANDLEY, D.M. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30(4):772-780, http://dx.doi.org/10.1631/jzus.B0860015
» http://dx.doi.org/10.1631/jzus.B0860015 - KUBICEK, C.P., KOMON-ZELAZOWSKA, M. & DRUZHININA, I.S. 2008. Fungal genus Hypocrea/ Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B 9(10):753-763, http://dx.doi.org/10.1631/jzus.B0860015
» http://dx.doi.org/10.1631/jzus.B0860015 - KULLNIG, C.M., SZAKACS, G. & KUBICEK, C.P. 2000. Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol Res 104:1117-1125, http://dx.doi.org/10.1017/S0953756200002604
» http://dx.doi.org/10.1017/S0953756200002604 - LOPES, R.B, BRITO, M.A.V., MELLO, S.C.M. & SAGGIN, O.J. 2013. Coleções microbianas na Embrapa: conservação e agregação de valor à biodiversidade, In: Simpósio Microrganismos em Agroenergia: da Prospecção aos Bioprocessos. Almeida JRM (Ed.). Brasília, DF: Embrapa Agroenergia (Documentos/Embrapa Agroenergia, ISSN 2177- 4439). p.15-22.
- MACHADO, D.F.M., PARZIANELLO, R., SILVA, A.C.F. & ANTONIOLLI, Z.I. 2012. Trichoderma no Brasil: O Fungo e o Bioagente. Ciências Agrárias 35(1):274-288.
- MATROUDI, I.S., ZAMANI, M.R. & MOTALLEBI, M. 2009. Antagonistic effects of three species of Trichoderma sp. on Sclerotinia sclerotiorum, the causal agent of canola stem rot. Egypt J Biol 11:37-44, http://dx.doi.org/10.4314/ejb.v11i1.56560
» http://dx.doi.org/10.4314/ejb.v11i1.56560 - MENTEN, J.O.M., MINUSSI, C.C., CASTRO, C. & KIMATI, H. 1976. Efeito de alguns fungicidas no crescimento micelial de Macrophomina phaseolina (Tass.) Goid. "in vitro". Fitopatol Bras 1(2):57-66, http://dx.doi.org/10.1590/S1983-40632013000400014
» http://dx.doi.org/10.1590/S1983-40632013000400014 - RAEDER, U. & BRODA, P. 1985. Rapid preparation of DNA from filamentous fungiLett Appl Microbiol 1(1):17-20, http://dx.doi.org/10.1111/j.1472-765X.1985.tb01479.x
» http://dx.doi.org/10.1111/j.1472-765X.1985.tb01479.x - RONQUIST, F. & HUELSENBECK, J.P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:72-4, http://dx.doi.org/10.1093/bioinformatics/btg180
» http://dx.doi.org/10.1093/bioinformatics/btg180 - SAMUELS, G.J., DODD, S.L., LU, B., PETRINI, O., SCHROERS, H. & DRUZHININA, I.S. 2006. The Trichoderma koningii aggregate species. Stud Mycol 56:67-133, http://dx.doi.org/10.3114/sim.2006.56.03
» http://dx.doi.org/10.3114/sim.2006.56.03 - SAMUELS, G.J., ISMAIEL, A., BOM, M.C., DE RESPINIS, S. & PETRINI, O. 2010. Trichoderma asperellum sensu lato consists of two crypitc species. Mycologia 104(4):944-966, http://dx.doi.org/10.3852/09-243
» http://dx.doi.org/10.3852/09-243 - WHITE, T.J., BRUNS, T., LEE, S. & TAYLOR, J.W. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, p.315-322, http://dx.doi.org/10.1016/B978-0-12-372180-8.50042-1
» http://dx.doi.org/10.1016/B978-0-12-372180-8.50042-1
Data availability
Data citations
AGROFIT. Busca por produtos formulados.http://agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons(last acess: 02/02/2016)
Publication Dates
-
Publication in this collection
2016
History
-
Received
20 June 2016 -
Reviewed
25 Aug 2016 -
Accepted
29 Aug 2016