Abstracts
The objective of this study was to design and evaluate new primers for species-specific detection of L. infantum chagasi using PCR. Two combinations of primer pairs were established with the aim of obtaining specific amplification products from the L. infantum chagasi 18S rRNA gene. The combinations of the primer pairs and the respective sizes of the PCR products, based on the U422465 GenBank reference sequence of L. infantum chagasi, were: LCS1/LCS3 (259 bp) and LCS2/LCS3 (820 bp). It was concluded that the new PCR assays optimized using the primer pairs LCS1/LCS3 and LCS2/LCS3 were effective for specific detection of L. infantum chagasi, with analytical sensitivity to detect 1 pg/µL of DNA.
Kala-azar; visceral leishmaniasis; molecular diagnosis; 18S rRNA gene; L. infantum chagasi
Objetivou-se com este trabalho construir e avaliar novos primers para a detecção espécie-específicos de L. infantum chagasi por PCR. Foram estabelecidas duas combinações de pares de primers com a finalidade de obter produtos de amplificação específicos para o gene 18S rRNA de L. infantum chagasi. As combinações dos pares de primers e os respectivos tamanhos dos produtos de PCR, previstos conforme a sequência de referência utilizada (GenBank U422465) foram: LCS1/LCS3 (259 pb); LCS2/LCS3 (820 pb). Pôde-se concluir que os novos ensaios de PCR otimizados neste estudo, empregando os pares de primers LCS1/LCS3 e LCS2/LCS3, foram efetivos para a detecção específica de L. infantum chagasi, com sensibilidade analítica para detectar 1 pg/µL de DNA.
Calazar; leishmaniose visceral; diagnóstico molecular; gene 18S rRNA; L. infantum chagasi
RESEARCH NOTE
Design of primer pairs for species-specific diagnosis of Leishmania (Leishmania) infantum chagasi using PCR
Construção de pares de primers para a detecção espécie-específica de Leishmania (Leishmania) infantum chagasi por PCR
Osvaldo José da Silveira Neto; Sabrina Castilho Duarte; Hérika Xavier da Costa; Guido Fontgalland Coelho Linhares
Laboratório de Doenças Parasitárias, Escola de Veterinária, Universidade Federal de Goiás - UFG, Goiânia, GO, Brasil
Corresponding author Corresponding author: Osvaldo José da Silveira Neto Laboratório de Doenças Parasitárias, Escola de Veterinária, Universidade Federal de Goiás - UFG, Campus II Goiânia, GO, Brasil e-mail: oswaldo_neto@hotmail.com
ABSTRACT
The objective of this study was to design and evaluate new primers for species-specific detection of L. infantum chagasi using PCR. Two combinations of primer pairs were established with the aim of obtaining specific amplification products from the L. infantum chagasi 18S rRNA gene. The combinations of the primer pairs and the respective sizes of the PCR products, based on the U422465 GenBank reference sequence of L. infantum chagasi, were: LCS1/LCS3 (259 bp) and LCS2/LCS3 (820 bp). It was concluded that the new PCR assays optimized using the primer pairs LCS1/LCS3 and LCS2/LCS3 were effective for specific detection of L. infantum chagasi, with analytical sensitivity to detect 1 pg/µL of DNA.
Keywords: Kala-azar, visceral leishmaniasis, molecular diagnosis, 18S rRNA gene, L. infantum chagasi.
RESUMO
Objetivou-se com este trabalho construir e avaliar novos primers para a detecção espécie-específicos de L. infantum chagasi por PCR. Foram estabelecidas duas combinações de pares de primers com a finalidade de obter produtos de amplificação específicos para o gene 18S rRNA de L. infantum chagasi. As combinações dos pares de primers e os respectivos tamanhos dos produtos de PCR, previstos conforme a sequência de referência utilizada (GenBank U422465) foram: LCS1/LCS3 (259 pb); LCS2/LCS3 (820 pb). Pôde-se concluir que os novos ensaios de PCR otimizados neste estudo, empregando os pares de primers LCS1/LCS3 e LCS2/LCS3, foram efetivos para a detecção específica de L. infantum chagasi, com sensibilidade analítica para detectar 1 pg/µL de DNA.
Palavras-chave: Calazar, leishmaniose visceral, diagnóstico molecular, gene 18S rRNA, L. infantum chagasi.
Introduction
Visceral leishmaniasis (VL) is a widespread zoonosis in tropical and subtropical regions of the planet, which causes high numbers of deaths, especially among untreated patients (BANETH, 2006; WHO, 2011). The species involved in VL infection are Leishmania donovani and Leishmania infantum chagasi (TAYLOR et al., 2010).
The conventional laboratory methods for diagnosing VL consist of serological techniques and direct parasitological examination. Among the serological tests, the indirect immunofluorescence assay (IFA) and the enzyme immunoassay (ELISA) are the ones most frequently used. Both show high sensitivity, although cross-reactions with other forms of leishmaniasis, Chagas disease and trypanosomiasis have been reported (GOMES et al., 2007; TRONCARELLI et al., 2008). Parasitological examination of either skin smears or aspirates from lymph nodes, spleen or bone marrow is useful for confirming the diagnosis, because of its high specificity.
Since 1998, the polymerase chain reaction (PCR) has been increasingly applied as an alternative method for diagnosing visceral leishmaniasis (TELLERIA et al., 1999; BRANDÃO-FILHO et al., 2003; CORTES et al. 2004; ANDRADE et al., 2006; GOMES et al., 2007; SALAM et al., 2010). Additional use of restriction enzymes on the kDNA-PCR product has been adapted by some researchers in order to demonstrate different fragment length polymorphisms (RFLPs) in the genus Leishmania (VOLPINI et al. 2004; ANDRADE et al., 2006). Because of the high sensitivity of the PCR technique, it is suitable for diagnostic purposes. The sensitivity and specificity of the PCR are directly related to the set of primers used for amplification of the target DNA, the number of copies of the target DNA to be amplified, the DNA extraction method used, the type of material to be analyzed and the PCR protocol (CORTES et al., 2004; SALAM et al., 2010).
Different amplification targets in the Leishmania genome have been reported. They most frequently correspond to repetitive and/or polymorphic sequence regions of the encoding genes for ribosomal RNAs, including internal transcribed spacers, β-tubulin, locus gp63, hsp70 and cysteine proteinase genes, as well as kinetoplast minicircle DNA (kDNA) (REITHINGER et al., 2000; SINGH, 2006). The latter has been considered to be one of the best targets for PCR (GONTIJO; MELO, 2004).
PCR has been successfully used to detect Leishmania DNA in several biological materials such as blood, bone marrow, skin, conjunctival samples and cerebrospinal fluid (LACHAUD et al., 2002; AOUN et al., 2009). Other advantages of PCR include the possibility of using small amounts of different clinical samples and detection of low concentrations of parasites (MELO, 2004).
The objective of the present study was to develop and evaluate new species- specific primers in order to discriminate L. infantum chagasi DNA using PCR.
Materials and Methods
Novel primers were designed from alignments of reference sequences for the 18S rRNA gene, from different species of Leishmania obtained from GenBank database (http://www.ncbi.nlm.nih.gov). For this procedure, the following sequences with the corresponding GenBank access numbers were used: L. infantum (U42465), L. infantum chagasi (GQ332359), L. donovani (GQ332356), L. guyanensis (GQ332358), L. amazonensis (GQ332354) and L. braziliensis (GQ332355).
The sequences were aligned by means of the Clustal W multiple progression method (THOMPSON et al., 2004), using the Megalign software (LaserGene, DNAStar, Inc.). The L. infantum chagasi-specific primers (sense and antisense) were then selected from non-conserved or semi-conserved regions of the gene. They were then subjected to similarity analysis against the GenBank database, using the BLAST-N® algorithm (Basic Local Alignment Search Tool - Nucleotides) (ALTSCHUL et al., 1990).
Preparation of the PCR reaction mix and amplification conditions was done by adapting the protocols described by Linhares et al. (2002) and optimizing them in accordance with Bartlett and Stirling (2003). The PCR was then carried out by using 2.5 µL of DNA template sample in a final volume of 50 µL of reaction mix composed of 20 mM Tris-HCl + 50 mM KCl (PCR buffer, Invitrogen), 20 mM MgCl2, 0.2 mM dNTP (Amersham Biosciences), 200 mM each primer, 1.5 U of Taq DNA polymerase (Invitrogen) and ultra-pure water (Dnase/Rnase-Free Distilled Water, Invitrogen).
The amplification reactions were conducted in a thermocycler (Mastercycler Personal), programmed for the following cycles and temperatures: initial denaturation step at 94 ºC for two minutes, followed by 40 repeated cycles with temperatures of 94 ºC for 30 seconds, 30 seconds at the annealing temperature calculated for each pair of primers, and one minute at 72 ºC, finishing the reaction with an additional extension phase at 72 ºC for two minutes.
The positive and negative controls were included in all reaction. The DNA controls used to assess the analytical sensitivity and specificity of the new primers were: genomic DNA of Canis familiaris, L. infantum chagasi, Leishmania braziliensis, Leishmania amazonensis, Leishmania guyanensis, Trypanosoma cruzi, Toxoplasma gondii, Babesia canis and Ehrlichia canis. Sterile ultrapure water, free of DNase and RNase (Dnase/Rnase-Free Distilled Water - Invitrogen), was used as the negative control.
The genomic DNA samples from Canis familiares, B. canis and E. canis used in this study were obtained from the Veterinary School, Federal University of Goiás (UFG). The samples of the genomic DNA of L. infantum chagasi were obtained from the Institute for Tropical Pathology and Public Health (IPTSP) of UFG and from São Paulo State University, Jaboticabal Campus. Samples of the genomic DNA of L. braziliensis (MHOM/BR/75/M2903), L. amazonensis (PH8 strain), L. guyanensis (PLR6), Trypanosoma cruzi (Y strain), and T. gondii (RH strain type I) were kindly provided by the Leishmaniasis Immunobiology Laboratory, IPTSP, UFG.
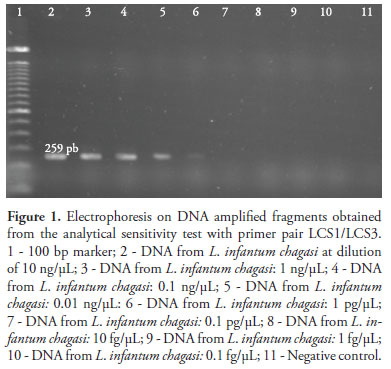
To investigate the analytical sensitivity, the genomic DNA of L. infantum chagasi was used with the initial concentration adjusted to 10 ng/µL. To establish the sensitivity threshold of the PCR assay, serial dilutions were prepared with the respective concentrations: 10 ng/µL, 1 ng/µL, 0.1 ng/µL, 0.01 ng/µL, 1 pg/µL, 0.1 pg/µL, 10 fg/µL, 1 fg/µL and 0.1 fg/µL.
A volume of 10 µL of the amplicons was applied to 1.2% agarose gel in 1X TBE buffer and was electrophoresed for 60 minutes at 90 V. The 100 bp DNA ladder (Invitrogen) was used as a molecular weight marker. The electrophoresis gels were stained with ethidium bromide solution (0.4 mg/mL) and the results were analyzed using a gel photodocumentation device (DocPrint, Vilber Lourmat) under UV illumination.
Results and Discussion
Two combinations of primer pairs were established to obtain specific amplification products from L. infantum chagasi. The names and combinations of primer pairs and the respective sizes of the expected PCR products, according to the reference sequence used (U422465), were as follows: LCS1/LCS3 (259 bp) and LCS2/LCS3 (820 bp).
The L. infantum chagasi-specific primers LCS1, LCS2 and LCS3 were selected from the alignment of 18S rRNA gene sequences of L. infantum chagasi, L. donovani, L. guyanensis, L. amazonensis and L. braziliensis, from a region with 100% similarity only to L. infantum chagasi (Table 1).
When used in PCR reactions with DNA samples from different species of the genus Leishmania, other microorganisms and C. familiaris, the primer pairs LCS1/LCS3 and LCS2/LCS3 generated amplification of the target fragments of 259 bp and 820 bp, respectively, only for L. infantum chagasi DNA. Nonspecific amplifications did not occur in any of the L. infantumchagasi specific assays.
In assessing the analytical sensitivity, PCR assays with both primer pairs, i.e. LCS1/LCS3 (Figure 1) and LCS2/LCS3 (Figure 2), presented the detection threshold of 1 pg/µL of L. infantum chagasi genomic DNA.
The detection threshold of 1 pg/µL of genomic DNA of L. infantum chagasi found in this study was less sensitive than seen in the results of Brujin and Barker (1992) and Roelfsema et al. (2011), who reported the threshold of 10 fg/µL, targeting DNA fragments of kinetoplast minicircles (kDNA) and the ITS region, respectively.
The greater sensitivity in amplification reactions using primers targeting kDNA of the Leishmania can be explained by the large number of copies of DNA present in the kinetoplast minicircle (REALE et al., 1999; LACHAUD et al., 2002). However, despite using primers for kDNA targets, Smyth et al. (1992) found the same sensitivity (1 pg/µL) reported in the present study. Although most authors have used the kinetoplast minicircle region for primer construction, the present study showed that primers selected from 18S rRNA are a reliable option for specific molecular detection of L. infantum chagasi.
Greater precision in analyzing the test sensitivity in comparison with the results presented by other authors is a difficult task to achieve, in two respects: (a) experiments conducted in different laboratories may be influenced by diverse factors, (b) information on the sensitivity threshold is neglected in most published papers.
Acknowledgements
To Dr. Miriam Cristina Leandro Dorta (Institute of Tropical Pathology and Public Health, UFG) for the samples of genomic DNA of L. braziliensis (MHOM/BR/75/M2903), L. amazonensis (PH8 strain), L. guyanensis (PLR6), Trypanosoma cruzi (Y strain) and T. gondii (RH strain type I). To Dr. Rosangela Zacarias Machado (UNESP, Jaboticabal, Brazil) for the L. infantum chagasi DNA.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215(3):403-410.
Andrade HM, Reis AB, Santos SL, Volpini AC, Marques MJ, Romanha AJ. Use of PCR-RFLP to identify Leishmania species in naturally-infected dogs. Vet Parasitol 2006;140(3-4):231-238. http://dx.doi.org/10.1016/j.vetpar.2006.03.031
Aoun O, Mary C, Roqueplo C, Marié JL, Terrier O, Levieuge A, et al. Canine leishmaniasis in south-east of France: Screening of Leishmania infantum antibodies (western blotting, ELISA) and parasitaemia levels by PCR quantification. Vet Parasitol 2009;166(1-2):27-31. http://dx.doi.org/10.1016/j.vetpar.2009.08.006
Baneth G. Leishmaniases. In: Greene CE. Infectious and Diseases of the dog and cat. Canada: Elsevier; 2006. p. 686-689.
Bartlett JMS, Stirling D. Methods in molecular biology: PCR Protocols. 2 ed. New Jersey: Humana Press; 2003. 519 p.
Brandão-Filho SP, Brito MEF, Carvalho FG, Ishikawa EA, Cupolillo E, Floeter-Winter LM, et al. Wild and synantropic hosts of Leishmania (Vianna) braziliensis in the endemic cutaneous leishmaniasis locality of Amaraji, Pernambuco State, Brazil. Trans Rev Soc Trop Méd Hyg 2003;97:291-296. http://dx.doi.org/10.1016/S0035-9203(03)90146-5
Brujin M, Barker DC. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop 1992;52(1):45-58. http://dx.doi.org/10.1016/0001-706X(92)90006-J
Cortes S, Rolão N, Ramada J, Campino L. PCR as a rapid and sensitive tool in the diagnosis of human and canine leishmaniasis using Leishmania donovani s. l.-specific kinetoplastid primers. Trans R Soc Trop Med Hyg 2004;98(1):12-17. http://dx.doi.org/10.1016/S0035-9203(03)00002-6
Gomes AH, Ferreira IM, Lima ML, Cunha EA, Garcia AS, Araújo MF, et al. PCR identification of Leishmania in diagnosis and control of canine Leishmaniasis. Vet Parasitol 2007;144(3-4):234-241. http://dx.doi.org/10.1016/j.vetpar.2006.10.008
Gontijo CM, Melo MN. Visceral Leishmaniasis in Brazil: current status, challenges and prospects. Rev Bras Epidemiol 2004;7(3):338-349. http://dx.doi.org/10.1590/S1415-790X2004000300011
Lachaud L, Hammami SM, Chabbert E, Dereure J, Dedet JP, Bastien P. Comparison of Six PCR Methods Using Peripheral Blood for Detection of Canine Visceral Leishmaniasis. J Clin Microbiol 2002;40(1):210-215. http://dx.doi.org/10.1128/JCM.40.1.210-215.2002
Linhares GFC, Santana AP, Laueman LH, Madruga CR. Assessment of primers designed from the small ribosomal subunit RNA for specific discrimination between Babesia bigemina and Babesia bovis by PCR. Ciênc Anim Bras 2002;3(2):27-32.
Melo MN. Leishmaniose visceral no Brasil: desafios e perspectivas. Rev Bras Parasitol Vet 2004;13(S1):41-45.
Reale S, Maxia L, Vitale F, Glorioso NS, Caracappa S, Vesco G. Detection of Leishmania infantum in dogs by PCR with lymph node aspirates and blood. J Clin Microbiol 1999;37(9):2931-2935.
Reithinger R, Lambson BE, Barker DC, Davies CR. Use of PCR to detect Leishmania (Viannia) spp. in dog blood and bone marrow. J Clin Microbiol 2000;38(2):748-751.
Roelfsema JH, Nozari N, Herremans T, Kortbeek LM, Pinelli E. Evaluation and improvement of two PCR targets in molecular typing of clinical samples of Leishmania patients. Exp Parasitol 2011;127(1):36-41. http://dx.doi.org/10.1016/j.exppara.2010.06.024
Salam MA, Mondal D, Kabir M, Ekram ARMS, Haque R. PCR for diagnosis and assessment of cure in kala-azar patients in Bangladesh. Acta Trop 2010;113(1):52-55. http://dx.doi.org/10.1016/j.actatropica.2009.09.005
Singh, S. New developments in diagnosis of leishmaniasis. Indian J Med Res 2006;123(3):311-330.
Smyth AJ, Ghosh A, Hassan MQ, Basu D, De Bruijn MHL, Adhya S, et al. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology 1992;105(Pt2):183-192. http://dx.doi.org/10.1017/S0031182000074096
Taylor MA, Coop RL, Wall RL. Parasitologia Veterinária. 3rd ed. Rio de Janeiro: Guanabara Koogan; 2010. 742 p.
Telleria J, Bosseno MF, Tarifa T, Buitrago R, Martinez E, Torrez M, et al. Putative reservoirs of Leishmania amazonensis in a Sub-andean focus of Bolivia identified by kDNA-Polymerase Chain Reaction. Mem Inst Oswaldo Cruz 1999;94(1):5-6. http://dx.doi.org/10.1590/S0074-02761999000100002
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 2004;22(22):4673-4680. http://dx.doi.org/10.1093/nar/22.22.4673
Troncarelli MZ, Machado JG, Camargo LB, Hoffmann JL, Camossi L, Greca H, et al. Associação entre resultados sorológicos no diagnóstico da leishmaniose e de tripanossomíase canina, pela técnica de imunofluorescência indireta. Vet e Zootec 2008;15:39-46.
Volpini AC, Passos VMA, Oliveira GC, Romanha AJ. PCR-RFLP to identify Leishmania (Viannia) braziliensis and L. (Leishmania) amazonensis causing American cutaneous leishmaniasis. Acta Trop 2004;90(1):31-37. http://dx.doi.org/10.1016/j.actatropica.2003.10.008
World Health Organization - WHO. Leishmaniasis: the global trend. Geneva, 2011. Available from http://www.who.int/neglected_diseases/integrated_media_leishmaniasis/.
Received September 8, 2011
Accepted August 23, 2012
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215(3):403-410.
- Andrade HM, Reis AB, Santos SL, Volpini AC, Marques MJ, Romanha AJ. Use of PCR-RFLP to identify Leishmania species in naturally-infected dogs. Vet Parasitol 2006;140(3-4):231-238. http://dx.doi.org/10.1016/j.vetpar.2006.03.031
- Aoun O, Mary C, Roqueplo C, Marié JL, Terrier O, Levieuge A, et al. Canine leishmaniasis in south-east of France: Screening of Leishmania infantum antibodies (western blotting, ELISA) and parasitaemia levels by PCR quantification. Vet Parasitol 2009;166(1-2):27-31. http://dx.doi.org/10.1016/j.vetpar.2009.08.006
- Baneth G. Leishmaniases. In: Greene CE. Infectious and Diseases of the dog and cat Canada: Elsevier; 2006. p. 686-689.
- Bartlett JMS, Stirling D. Methods in molecular biology: PCR Protocols. 2 ed. New Jersey: Humana Press; 2003. 519 p.
- Brandão-Filho SP, Brito MEF, Carvalho FG, Ishikawa EA, Cupolillo E, Floeter-Winter LM, et al. Wild and synantropic hosts of Leishmania (Vianna) braziliensis in the endemic cutaneous leishmaniasis locality of Amaraji, Pernambuco State, Brazil. Trans Rev Soc Trop Méd Hyg 2003;97:291-296. http://dx.doi.org/10.1016/S0035-9203(03)90146-5
- Brujin M, Barker DC. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop 1992;52(1):45-58. http://dx.doi.org/10.1016/0001-706X(92)90006-J
- Cortes S, Rolão N, Ramada J, Campino L. PCR as a rapid and sensitive tool in the diagnosis of human and canine leishmaniasis using Leishmania donovani s. l-specific kinetoplastid primers. Trans R Soc Trop Med Hyg 2004;98(1):12-17. http://dx.doi.org/10.1016/S0035-9203(03)00002-6
- Gomes AH, Ferreira IM, Lima ML, Cunha EA, Garcia AS, Araújo MF, et al. PCR identification of Leishmania in diagnosis and control of canine Leishmaniasis. Vet Parasitol 2007;144(3-4):234-241. http://dx.doi.org/10.1016/j.vetpar.2006.10.008
- Gontijo CM, Melo MN. Visceral Leishmaniasis in Brazil: current status, challenges and prospects. Rev Bras Epidemiol 2004;7(3):338-349. http://dx.doi.org/10.1590/S1415-790X2004000300011
- Lachaud L, Hammami SM, Chabbert E, Dereure J, Dedet JP, Bastien P. Comparison of Six PCR Methods Using Peripheral Blood for Detection of Canine Visceral Leishmaniasis. J Clin Microbiol 2002;40(1):210-215. http://dx.doi.org/10.1128/JCM.40.1.210-215.2002
- Melo MN. Leishmaniose visceral no Brasil: desafios e perspectivas. Rev Bras Parasitol Vet 2004;13(S1):41-45.
- Reale S, Maxia L, Vitale F, Glorioso NS, Caracappa S, Vesco G. Detection of Leishmania infantum in dogs by PCR with lymph node aspirates and blood. J Clin Microbiol 1999;37(9):2931-2935.
- Reithinger R, Lambson BE, Barker DC, Davies CR. Use of PCR to detect Leishmania (Viannia) spp. in dog blood and bone marrow. J Clin Microbiol 2000;38(2):748-751.
- Roelfsema JH, Nozari N, Herremans T, Kortbeek LM, Pinelli E. Evaluation and improvement of two PCR targets in molecular typing of clinical samples of Leishmania patients. Exp Parasitol 2011;127(1):36-41. http://dx.doi.org/10.1016/j.exppara.2010.06.024
- Salam MA, Mondal D, Kabir M, Ekram ARMS, Haque R. PCR for diagnosis and assessment of cure in kala-azar patients in Bangladesh. Acta Trop 2010;113(1):52-55. http://dx.doi.org/10.1016/j.actatropica.2009.09.005
- Singh, S. New developments in diagnosis of leishmaniasis. Indian J Med Res 2006;123(3):311-330.
- Smyth AJ, Ghosh A, Hassan MQ, Basu D, De Bruijn MHL, Adhya S, et al. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology 1992;105(Pt2):183-192. http://dx.doi.org/10.1017/S0031182000074096
- Taylor MA, Coop RL, Wall RL. Parasitologia Veterinária 3rd ed. Rio de Janeiro: Guanabara Koogan; 2010. 742 p.
- Telleria J, Bosseno MF, Tarifa T, Buitrago R, Martinez E, Torrez M, et al. Putative reservoirs of Leishmania amazonensis in a Sub-andean focus of Bolivia identified by kDNA-Polymerase Chain Reaction. Mem Inst Oswaldo Cruz 1999;94(1):5-6. http://dx.doi.org/10.1590/S0074-02761999000100002
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 2004;22(22):4673-4680. http://dx.doi.org/10.1093/nar/22.22.4673
- Troncarelli MZ, Machado JG, Camargo LB, Hoffmann JL, Camossi L, Greca H, et al. Associação entre resultados sorológicos no diagnóstico da leishmaniose e de tripanossomíase canina, pela técnica de imunofluorescência indireta. Vet e Zootec 2008;15:39-46.
- Volpini AC, Passos VMA, Oliveira GC, Romanha AJ. PCR-RFLP to identify Leishmania (Viannia) braziliensis and L. (Leishmania) amazonensis causing American cutaneous leishmaniasis. Acta Trop 2004;90(1):31-37. http://dx.doi.org/10.1016/j.actatropica.2003.10.008
- World Health Organization - WHO. Leishmaniasis: the global trend. Geneva, 2011. Available from http://www.who.int/neglected_diseases/integrated_media_leishmaniasis/
Corresponding author:
Publication Dates
-
Publication in this collection
11 Oct 2012 -
Date of issue
Sept 2012
History
-
Received
08 Sept 2011 -
Accepted
23 Aug 2012