ABSTRACT
The selection of superior genotypes of soybean is a complex process, thus exploratory multivariate techniques can be applied to select genotypes analyzing the agronomic traits altogether, increasing the chance of success of a breeding program. Thus, the objective of this study was to select soybean genotypes carrying the RR gene with good agronomical performance through of multivariate analysis and selection index and identify those traits that influence, also verifying the agreement of multivariate techniques and selection index in the selection process. The experiment was conducted in an increased block experimental being evaluated 227 genotypes of F5 generation, which 85 of those were detected to be glyphosate-resistant by PCR. The following traits were evaluated: number of days to maturity, plant height at maturity, lodging, agronomic value, number of branches per plant, number of pods per plant, hundred seeds weight and grain yield. The principal components analysis resulted in the selection of sixteen genotypes with higher grain yield. The traits related to the production of components exerted great influence on grain yield. The clustering by K-means and Ward's methods were similar because they clustered the specific genotypes for the selected traits in the principal components analysis in the same group. There was an agreement on the results of the multivariate analysis in the selection index of Mulamba and Mock in relation to the selected genotypes. The methodologies applied are efficient for selecting genotypes.
Key words:
Glycine max; Clustering analysis; Principal components; Selection gain; Grain Yield
RESUMO
A seleção de genótipos superiores de soja é um processo complexo; dessa forma, técnicas exploratórias multivariadas podem ser aplicadas para selecionar genótipos, analisando simultaneamente todos os caracteres agronômicos em estudo, aumentando a chance de êxito em um programa de melhoramento. O objetivo do presente estudo consistiu em selecionar genótipos de soja portadores do gene RR com bons caracteres agronômicos por meio de análise multivariada e índice de seleção, e identificar quais caracteres mais influenciam, além de verificar a concordância das técnicas multivariadas e índice de seleção no processo seletivo. O experimento foi conduzido no delineamento experimental de blocos aumentados, sendo avaliados 227 genótipos de soja de geração F5, sendo 85 destes, detectados pela técnica de PCR, como resistentes ao Roundup. Os caracteres avaliados foram: número de dias para maturidade, altura da planta na maturidade, acamamento, valor agronômico, número de ramos por planta, número de vagens por planta, peso de cem sementes e produtividade de grãos. A análise de componentes principais auxiliou na seleção de dezesseis genótipos, com maior produtividade de grãos. Os caracteres relacionados aos componentes de produção exerceram grande influência sobre a produtividade de grãos. Os resultados das análises de agrupamento pelo método de K-means e pelo o método de Ward quando comparados foram semelhantes, pois agruparam os genótipos específicos para os caracteres selecionados na análise de componentes principais, em um mesmo grupo. Houve concordância dos resultados das análises multivariadas com o índice de seleção de Mulamba e Mock quanto aos genótipos selecionados. As metodologias aplicadas são eficientes no processo de seleção de genótipos.
Palavras-chave:
Glycine max; Análise de agrupamento; Componentes principais; Ganho de seleção; Produtividade de grãos
INTRODUCTION
The soybean culture is a very important part of Brazilian agriculture. In 2016/17, Brazil produced 113.92 million tons of soybean (CONAB, 2017COMPANHIA NACIONAL DE ABASTECIMENTO. Acompanhamento da safra brasileira de grãos: 9º levantamento grãos safra 2016/17. Junho 2017. Available at: <http://www.conab.gov.br/OlalaCMS/uploads/arquivos/17_06_08_09_02_48_boletim_graos_junho_2017.pdf>. Accessed on July 07, 2017.
http://www.conab.gov.br/OlalaCMS/uploads...
) and the areas seeded with genetically modified cultivars reached 96.51% of total culture area, corresponding to 32.7 million hectares (CÉLERES, 2016CÉLERES. Acompanhamento da adoção de biotecnologia agrícola no Brasil: 2º levantamento grãos safra 2016/17. Dezembro 2016. Available at: <http://www.celeres.com.br/2o-levantamento-de-adocao-da-biotecnologia-agricola-no-brasil-safra-201617/. Accessed on July 07, 2017.
http://www.celeres.com.br/2o-levantament...
). The emergence of soybean cultivars resistant to the Roundup herbicide significantly changed weed management in these crops by providing benefits, such as efficient control and management flexibility in the post-emergence stage (MATSUO et al., 2009MATSUO, E. et al. Resistance of soybean genotypes to glyphosate. Planta Daninha, v. 27, p. 1063-1073, 2009. Número especial.; MENEGATTI; BARROS, 2007MENEGATTI, A. L. A.; BARROS, A. L. M. D. Análise comparativa dos custos de produção entre soja transgênica e convencional: um estudo de caso para o Estado do Mato Grosso do Sul. Revista de Economia e Sociologia Rural, v. 45, n. 1, p. 163-183, 2007.).
Selecting superior soybean genotypes is a complex process because the economically important agronomic traits are quantitative by nature and are correlated to each other (ALMEIDA; PELUZIO; AFFERRI, 2010ALMEIDA, R. D.; PELUZIO, J. M.; AFFERRI, F. S. Correlações fenotípicas, genotípicas e ambientais em soja cultivada sob condições várzea irrigada, sul do Tocantins. Bioscience Journal, v. 26, n. 1, p. 95-99, 2010.; NOGUEIRA et al., 2012NOGUEIRA, A. P. O. et al. Análise de trilha e correlações entre caracteres em soja cultivada em duas épocas de semeadura. Bioscience Journal, v. 28, n. 6, p. 877-888, 2012.). Promising genotypes should simultaneously unite a set of positive traits, which could elevate yield to fulfill the market demand (CRUZ, 2013CRUZ, C. D. Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum. Agronomy, v. 35, n. 3, p. 271-276, 2013.). Taking into consideration the complexity of the most important plant traits, we believe more efficient selection criteria is required.
Multivariate exploratory techniques can be used for selecting superior genotypes, simultaneously analyzing all the agronomic traits and the relationships they have with each other, as well as discriminating which is the most influencing in a selection process. Among the types of multivariate techniques, the analyses that stand out are the principal components analysis, and the hierarchical and non-hierarchical cluster analysis methods (HAIR et al., 2009HAIR, J. F. et al. Análise multivariada de dados. 6. ed. Porto Alegre: Bookman, 2009. 688 p.).
The main components analysis aims to reduce dataset variability, keeping as much relevant information as possible in the lowest number of components (BARBOSA et al., 2013BARBOSA, R. M., et al. Discrimination of soybean seed lots by multivariate exploratory techniques. Journal of Seed Science, v. 35, n. 3, p. 302-310, 2013.; DALLASTRA et al., 2014DALLASTRA, A. et al. Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014.; SILVA et al., 2010SILVA, M. O. et al. Discriminação de diferentes classes de solos irrigados com águas salinas, na região de Mossoró (RN), com o uso de análise multivariada. Ambiência, v. 6, n. 2, p. 261-270, 2010.). The cluster analysis, on the other hand, aims to classify similar individuals into the same group, while forming a heterogeneous set of groups (VIANNA et al., 2013VIANNA, V. F. et al. The multivariate approach and influence of characters in selecting superior soybean genotypes. African Journal of Agricultural Research, v. 8, p. 4162-4169, 2013.). Genotype selection studies have been performed with these techniques on several different cultures, according to Bertini et al. (2010)BERTINI, C. H. C. M. et al. Análise multivariada e índice de seleção na identificação de genótipos superiores de feijão-caupi. Acta Scientiarum. Agronomy, v. 32, n. 4, p. 613-619, 2010.; Dallastra et al. (2014)DALLASTRA, A. et al. Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014. and Vianna et al. (2013)VIANNA, V. F. et al. The multivariate approach and influence of characters in selecting superior soybean genotypes. African Journal of Agricultural Research, v. 8, p. 4162-4169, 2013.. In addition, multivariate methods have also been used in soybean genetic divergence studies (FERREIRA et al., 2015FERREIRA, J. A. et al. Diversidade genética em linhagens avançadas de soja oriundas de cruzamentos biparentais, quádruplos e óctuplos. Revista Ciência Agronômica, v. 46, n. 2, p. 339-351, 2015.; PELUZIO et al., 2012PELUZIO, J. M. et al. Genetic divergence among soybean cultivars in irrigated lowland in the State of Tocantins. Ciência Rural, v. 42, n. 3, p. 395-400, 2012.; VILLELA et al., 2014VILLELA, O. et al. Genetic divergence of roundup ready (RR) soybean cultivars estimated by phenotypic characteristics and molecular markers. African Journal of Biotechnology, v. 13, n. 26, p. 2613-2625, 2014.).
Selection indexes are methodologies that can identify superior genotypes. They enable researchers to efficiently select a set of economically expressive agronomic traits, increasing the success of the genetic improvement program (CRUZ, 2013CRUZ, C. D. Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum. Agronomy, v. 35, n. 3, p. 271-276, 2013.; ROSADO et al., 2012ROSADO, L. D. S. et al. Simultaneous selection in progenies of yellow passion fruit using selection indices. Revista Ceres, v. 59, n. 1, p. 95-101, 2012.).
The use of indexes to select soybean genotypes can promote better total gains, distributed among all the available traits, which is adequate for genetic improvement programs (COSTA et al., 2004COSTA, M. M. et al. Ganho genético por diferentes critérios de seleção em populações segregantes de soja. Pesquisa Agropecuária Brasileira, v. 39, n. 11, p. 1095-1102, 2004.). Selection indexes were used in recent soybean-traits studies (BIZARI et al., 2017BIZARI, E. H. et al. Selection indices for agronomic traits in segregating populations of soybean. Revista Ciência Agronômica, v. 48, n. 1, p. 110-117, 2017.; LEITE et al., 2016LEITE, W. S. et al. Estimativas de parâmetros genéticos, correlações e índices de seleção para seis caracteres agronômicos em linhagens F8 de soja. Comunicata Scientiae, v. 7, n. 3, p. 302-310, 2016.). Notably, the Mulamba and Mock (1978)MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978. sum of ranks index consists of classifying the genotypes according to each trait in favorable order of improvement and has the advantage of not needing to establish economic importance and variance and covariance estimations (REZENDE et al., 2014REZENDE, J. C. et al. Genetic progress in coffee progenies by different selection criteria. Coffee Science, v. 9, n. 3, p. 347-353, 2014.).
Based on the aforementioned considerations, the present study aims to select Roundup Ready soybean genotypes with positive agronomic traits, using multivariate assessments and selection indexes. Furthermore, it intends to identify which traits influence selection most and evaluate the relationship between these features, as well as verify the correlations between multivariate techniques and selection indexes in a selective process.
MATERIAL AND METHODS
Agronomic analyses
The experiment was conducted in the 2013/2014 agricultural year at the Jaboticabal Campus of UNESP/FCAV (São Paulo State University), located in the northern region of São Paulo State (21°15'22" S latitude and 48°18'58" W longitude). The genotypes evaluated in this study were obtained by artificial hybridization between conventional genitors (seed-bearing parent), lineages of the FCAV/UNESP Jaboticabal genetic improvement program, and genitors with the RR gene, commercial cultivars (male parent). In total, twenty genetic crossing were assessed.
Treatments consisted of 227 F5 generation soybean lineages, regardless of having the RR gene or not, since up to the present generation they had not been selected for glyphosate resistance. The augmented block design proposed by Federer (1956)FEDERER, W. T. Augmented (or hoonuiaku) designs. Hawaiian Planters Record, v. 55, n. 1, p. 191-208, 1956. was used, in which the plot was composed by a 5-meter long row with 0.5-meter spacing. Two check cultivars were used as interspersed controls (CD-216 and Vmax).
The following agronomic traits were assessed: number of days to maturity (NDM); plant height at maturity (PHM) in cm; lodging (L) visual scoring system, varying from 1 (all plot plants erect) to 5 (all plot plants lodged); agronomic value (AV) visual scoring system, varying from 1 (unwanted plants) to 5 (good plants); number of branches per plant (NB); number of pods per plant (NP); weight of 100 seeds (WHS) in g; and grain yield (GY) converted into kg ha-1.
Molecular analyses
The Roundup Ready (RR) soybean genotypes were detected by the PCR laboratory technique, which distinguishes genotypes through the presence of molecular imprints, resulting from specific transgenic DNA sequences, which are amplified. Genotypes containing the RR gene were identified through specific oligonucleotides that pair with the Forward sequence 5'TGATGTGATATCTCCACTGACG 3' and Reverse 5'TGTATCCCTTGAGCCATGTTGT 3', which encodes the EPSPS region. The RR sequence has a fragment size of 172pb (MARCELINO; GUIMARÃES; BARROS, 2007MARCELINO, F. C.; GUIMARÃES, M. F. M.; BARROS, E. G. Detecção e quantificação de alimentos geneticamente modificados: o panorama brasileiro. Revista Ceres, v. 54, n. 313, p. 239-249, 2007.).
The genomic DNA samples were extracted from the tissue of young trefoil, using the CTAB method described by Ferreira and Grattapaglia (1995)FERREIRA, M. E.; GRATTAPAGLIA, D. Introdução ao uso de marcadores RAPD e RFLP em análise genética. Brasília: Emprapa-Cenargen, 1995. 220 p. (EMBRAPA-CENARGEN. Documentos, 20).. The PCR reactions and electrophoresis separation of the amplified fragments in agarose gels were conducted according to Silva et al. (2015)SILVA, F. M. et al. Toothpick test: a methodology for the detection of RR soybean plants. Revista Ciência Agronômica, v. 46, n. 2, p. 436-442, 2015..
Table 1 exhibits a list of the crossings and genitors used to form the segregating populations, as well as the number of soybean genitors that have the RR gene, characterized and identified in generation F5 for each crossing. Among all the assessed genotypes, both crossing number 14 and 15 presented no RR gene in their genotypes.
Crossings and respective genotypes, the total number of genotypes, number of identified RR genotypes and their codes as a function of the number of identified genotypes per crossing
Statistical analyses
The soybean genotypes containing the RR gene were encoded from 1 to 85. Further selection was conducted based on the assessed agronomic traits.
Data were standardized and resulted in an average of zero and a variance of one for all variables as recommended in multivariate analysis (STATSOFT, 2004STATSOFT. Statistica: data analysis software system, version 10. Tulsa, 2004. Available at: <http://www.statsoft.com>. Accessed on July 25, 2015.
http://www.statsoft.com...
). Given the dependence structure and complexity of the original set of variables, multivariate analyses of principal components, Ward's hierarchical clustering, and K-means non-hierarchical clustering were performed for RR genotypes. These analyses were to improve the discrimination of superior genotypes and compare the results from exploratory analyses, as well as to test the correlation of the results from all the applied methods. All multivariate statistical analyses were performed using the STATISTICA v.10 software (STATSOFT, 2004STATSOFT. Statistica: data analysis software system, version 10. Tulsa, 2004. Available at: <http://www.statsoft.com>. Accessed on July 25, 2015.
http://www.statsoft.com...
).
Genotypes were selected by the principal components analyses, establishing criteria in which the scales of the graphs were pre-determined through ellipse on the graph axes to enable precise discrimination of progenies based on their different traits, according to Dallastra et al. (2014)DALLASTRA, A. et al. Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014. and Vianna et al. (2013)VIANNA, V. F. et al. The multivariate approach and influence of characters in selecting superior soybean genotypes. African Journal of Agricultural Research, v. 8, p. 4162-4169, 2013.. The lowest scale is the least stringent and consists of an X-axis varying from 2 to -2 and Y axis from 2 to -2. The second scale, more complex, varies on the X axis from 3 to -3 and on the Y axis from 3 to -3. This pattern goes on for all biplot graphs. Therefore, Plant breeders have to decide on the best level of selection intensity to use in the analysis and, hence, in the number of individuals selected for the interesting traits.
The Ward's clustering generated a dendrogram to obtain prior information about the number of groups that would be formed by the set of genotypes, based on the established between-group linkage distance. The dissimilarity measurement between the genotypes was based on the Euclidian distance (HAIR et al., 2009HAIR, J. F. et al. Análise multivariada de dados. 6. ed. Porto Alegre: Bookman, 2009. 688 p.).
In the K-means cluster analysis, we predetermined that 5 groups of RR soybean genotypes would be formed. The observation on the graphs indicate the "centers" of the groups for the analyzed traits. To determine the number of groups, the prior information created on the dendrogram was considered, based on the number of groups formed according to the adopted group connection distance. Therefore, with the K-means , we aimed to minimize the distance between each point and its centroid (HAIR et al., 2009HAIR, J. F. et al. Análise multivariada de dados. 6. ed. Porto Alegre: Bookman, 2009. 688 p.).
For the RR soybean genotypes, the sum of ranks selection index, described by Mulamba and Mock (1978)MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978. with 19% intensity, was used, aiming to select the same number of genotypes discriminated by the main components analysis for results comparisons. This analysis was processed by Genes - genetic software (CRUZ, 2013CRUZ, C. D. Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum. Agronomy, v. 35, n. 3, p. 271-276, 2013.).
RESULTS AND DISCUSSION
In the principal components analysis, three eigenvalues ware higher than one, explaining 67.58% of the variance contained in the eight original variables. Eigenvalues above one (1.0) generate components with significant quantities of information of the original variables (BERTINI et al., 2010BERTINI, C. H. C. M. et al. Análise multivariada e índice de seleção na identificação de genótipos superiores de feijão-caupi. Acta Scientiarum. Agronomy, v. 32, n. 4, p. 613-619, 2010.; DALLASTRA et al., 2014DALLASTRA, A. et al. Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014.). However, for those below one, the quantity of information retained in the component has no relevance.
The eigenvalue for the first principal component (PC1) was 2.47, which corresponds to 30.92% of total variance. The main variables that explained the variance retention were PHM, L, and AV, with correlation values of PC1 above 0.5 (Table 2), since the values above this magnitude, regardless of the signal, contribute the most to the generated component. The eigenvalue for the second main component (PC2) was 1.81, which retained 22.63% of the total variance, explained especially by the traits NB, NP, and GY. The third main component (PC3) retained 14.03% of the variance, explained mainly by the NB variable. For each principal component, same signals mean that the traits are positively correlated, and opposite signals mean the traits are negatively correlated (HAIR et al., 2009HAIR, J. F. et al. Análise multivariada de dados. 6. ed. Porto Alegre: Bookman, 2009. 688 p.).
Correlation between the agronomic traits and the principal components (PC) of the 85 soybean genotypes with the RR gene in generation F5
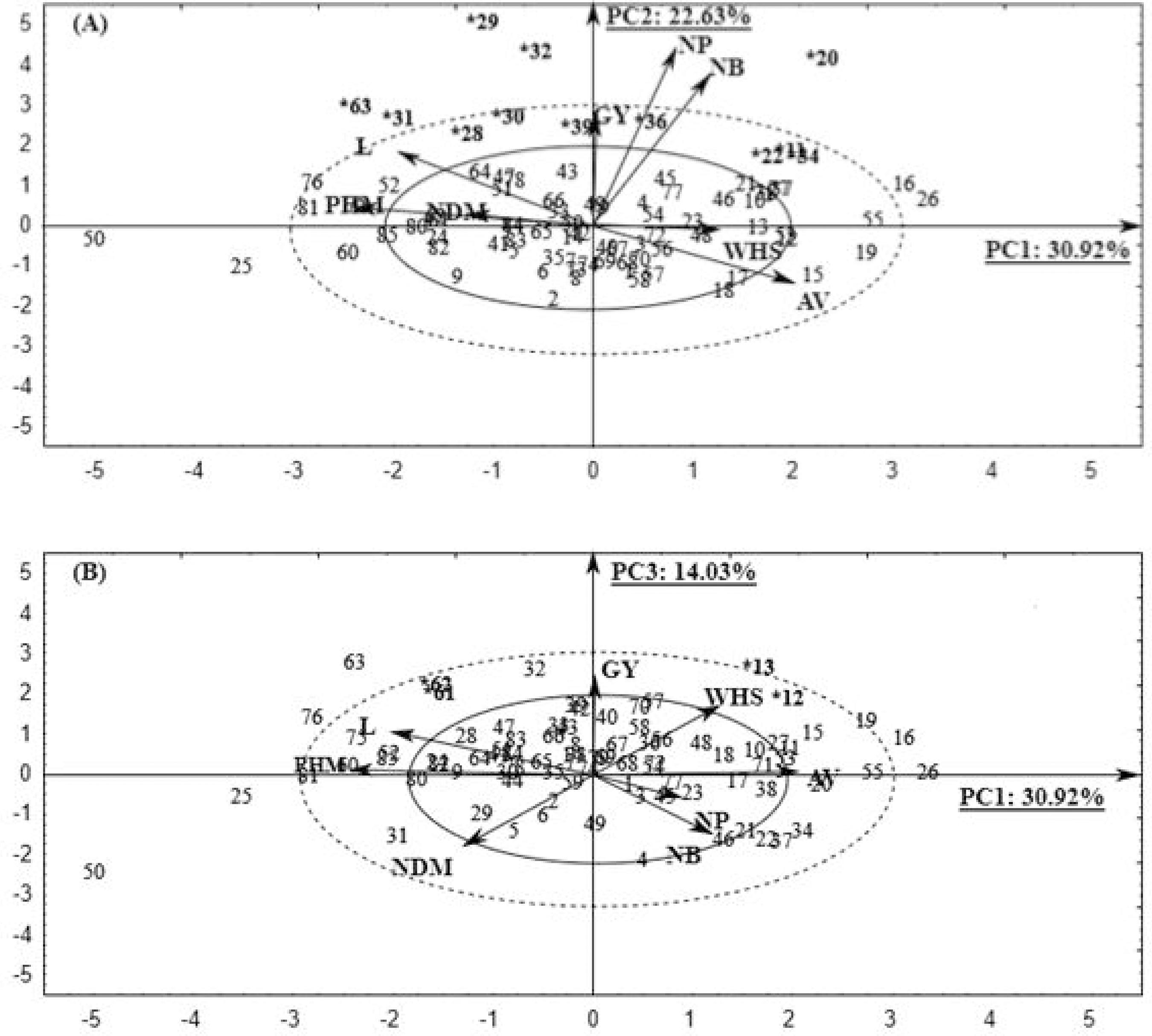
The two-dimensional plane, formed by PC1 (30.92%) and PC2 (22.63%), retained a total of 53.55% of the original variance (Figure 1A), which was the highest variance when correlating the most relevant components. This variance is explained by the following traits: PHM, L, AV, NB, NP, and GY and enabled us to choose the specific genotypes 11; 22;28;30;31;34;36; and 63; and the highly specific characters 20; 29; and 32 for the traits NB, NP, and GY. The highly specific genotypes are those that are well discriminated by one or a few traits and are highly superior since each genotype trait exhibits high influence. The specific genotypes, on the other hand, are those discriminated by several traits. The two-dimensional plane formed by the PC1 (30.92%) and PC3 (14.03%) retained 44.95% of remaining variance (Figure 1B) and the genotypes 12, 13, 61, and 62 were selected and discriminated by the traits GY, WHS, NDM, PHM, and L. Genotypes 12 and 13 exhibited superior GY and WHS, as well as inferior NDM, PHM, and L results, which are important and benign to the selection. Genotypes 61 and 62 exhibited superior GY and PHM, but exhibited and a higher level of L, indicating the intense relationship between PHM and L.
Biplot graph with 85 RR soybean genotypes dispersed in function to the principal components: PC1 x PC2 (A) and PC1 x PC3 (B) and projection of the traits vectors: NDM - Number of day to maturity (days); PHM - Plant height at maturity (cm), L - Lodging (visual score); AV - Agronomic value (visual score); NB - Number of branches (un); NP - Number of pods (un); WHS - Weight of 100 seeds (g) and GY - Grain yield (kg ha-1)
A large group of genotypes was positioned at the center of the graphs, meaning that they exhibit mean values for all the traits, which hinders identification of the more influential traits. Therefore, the trait values are within the range that classifies them as regular (not superior), which also exhibits the homogeneity of that group.
The genotypes located within the established scales are specific for the agronomic traits that influence them (Figure 1A and 1B). On the other hand, regarding the genotypes located in the graph extremities, they exhibit higher performance than the others in some specific traits since they possess very specific traits. Therefore, genotypes 20, 29, and 32 have extremely specific standards for the traits NB, NP, and GY because of their superior phenotypic values.
Figure 1A exhibits dispersion of the traits vectors according to the size proportion of the vectors among each other. Concerning the selected genotypes, the NB and NP traits exhibit a highly positive correlation with GY and are positioned in the same graph quadrant. These results are in agreement with those obtained in other studies (NOGUEIRA et al., 2012NOGUEIRA, A. P. O. et al. Análise de trilha e correlações entre caracteres em soja cultivada em duas épocas de semeadura. Bioscience Journal, v. 28, n. 6, p. 877-888, 2012.; VIANNA et al., 2013VIANNA, V. F. et al. The multivariate approach and influence of characters in selecting superior soybean genotypes. African Journal of Agricultural Research, v. 8, p. 4162-4169, 2013.). Thus, such traits compose the yield components, i.e., the larger their values the higher the soybean yield of that genotype, which facilitates the selection of superior genotypes with better efficiency. According to the projections of the GY, NDM, PHM, and L traits vectors (Figure 1B), genotypes 12 and 13, besides having the highest soybean yield, also exhibit better precocity, smaller size, and non-lodging shape.
Because of the number of pods, mature plant height is a trait that has a great indirect effect on the productivity of the soybeans. Thus, this trait can be used in genetic enhancement for indirectly selecting soybean yield (ALCANTARA NETO et al., 2011ALCANTARA NETO, F. et al. Análise de trilha do rendimento de grãos de soja na microrregião do Alto Médio Gurguéia. Comunicata Scientiae, v. 2, n. 2, p. 107-112, 2011.; ALMEIDA; PELUZIO; AFFERRI, 2010ALMEIDA, R. D.; PELUZIO, J. M.; AFFERRI, F. S. Correlações fenotípicas, genotípicas e ambientais em soja cultivada sob condições várzea irrigada, sul do Tocantins. Bioscience Journal, v. 26, n. 1, p. 95-99, 2010.).
Figure 1B shows that the WHS trait was most important to good soybean yield since both are strongly associated and positioned in the same graph quadrant. This can be explained by the fact that the genotypes present higher WHS when the size of the seeds are larger, because of the smaller quantity produced by the plant, since these genotypes suffered almost no influence from the yield components NB and NP. According to Dallastra et al. (2014)DALLASTRA, A. et al. Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014., the lower the number of pods the lower the number of beans per plant and higher the weight of 100 beans.
Concerning the group of selected genotypes, not all crossings were represented. Therefore, analyzing the parents involved in the crossing, we observed that the female genitors JAB.00-05-5/4A2D and JAB.00-06-2/3I3D were more efficient, with 6 and 5 selected RR genotypes, respectively. Regarding male genitors, M 8230 RR was most effective, with 5 selected genotypes. According to Gonçalves et al. (2014)GONÇALVES, A. et al. Estimating combining ability in popcorn lines using multivariate analysis. Chilean Journal of Agricultural Research, v. 74, n. 1, p. 10-15, 2014., in order to form segregating populations, selecting the genitors is a crucial step in guaranteeing success in genetic enhancement programs, in which the combination capacity with the presence of complementary genes is largely responsible for the success.
Figure 2 exhibits the dendrogram generated by the Ward cluster analysis. In general, it shows two large groups, named groups A and B. In a rougher selection, through the adopted cutoff point, five subgroups were formed, two of which were within Group A (A1 and A2) and three within group B (B1, B2, and B3). In regard to the subgroups, the genetic difference between the genotypes of these populations was verified based on the studied traits. However, individuals of the same subgroup were considered very similar, while those from different subgroups were considered very distinct.
Dendrogram representing the group of 85 RR soybean genotypes, obtained by the Ward method from 8 agronomic traits
In subgroup A2, only the genotypes selected in the principal components analysis were classified, since they have similar grain yield traits, number of branches and number of pods. The two genotypes selected by main component analysis were grouped in subgroup B1, representing genotypes with greater precocity, besides showing a great influence of a 100-grain variable on crop yield. This is because these genotypes present high similarity for these traits.
Genotypes 28, 61, and 62, from subgroup A1, exhibited similarly high soybean yield as well as mature plant height, and, consequently, elevated lodging rate. On the other hand, genotypes 11 and 34 from subgroups B2 presented similarly low mature plant heights, low lodging rates, and high soybean yield.
Figure 3 exhibits the cluster graph analysis generated by the K-means method, with 85 soybean genotypes for eight agronomic traits, distributed into five groups (clusters). The classification of genotypes is listed in Table 3.
Line graph of each group's centroid profiles by the K-means method and Euclidian distance from the agronomic traits: NDM - Number of day to maturity (days); PHM - Plant height at maturity (cm), L - Lodging (visual score); AV - Agronomic value (visual score); NB - Number of branches (un); NP - Number of pods (un); WHS - Weight of 100 seeds (g) and GY - Grain yield (kg ha-1) of 85 RR soybean genotypes
Optimization grouping between 85 RR soybean genotypes, obtained by the K-means method, based on 8 agronomic traits, using the Euclidian distance
Group 4 (Figure 3) was characterized as above average for NDM, PHM, and L, and below average for agronomic values. Furthermore, significantly high values of NB and NP were found for this same group. Thus, the genotypes placed in this group presented the highest GY results from all the groups formed in this analysis. Therefore, this reemphasizes that the NB and NP combination directly determines the yield potential of a soybean genotype. Highly positive phenotypic correlation results between these two traits were obtained in other studies (ALCANTARA NETO et al., 2011ALCANTARA NETO, F. et al. Análise de trilha do rendimento de grãos de soja na microrregião do Alto Médio Gurguéia. Comunicata Scientiae, v. 2, n. 2, p. 107-112, 2011.; ALMEIDA; PELUZIO; AFFERRI, 2010ALMEIDA, R. D.; PELUZIO, J. M.; AFFERRI, F. S. Correlações fenotípicas, genotípicas e ambientais em soja cultivada sob condições várzea irrigada, sul do Tocantins. Bioscience Journal, v. 26, n. 1, p. 95-99, 2010.; LEITE et al., 2016LEITE, W. S. et al. Estimativas de parâmetros genéticos, correlações e índices de seleção para seis caracteres agronômicos em linhagens F8 de soja. Comunicata Scientiae, v. 7, n. 3, p. 302-310, 2016.).
Group 5 (Figure 3) was characterized by exhibiting genotypes with values significantly lower than average for NDM, PHM, L, AV, NP, and GY. This genotype group presented the highest genotype precocity and reduced mature plant height, however, when compared to the other groups, its yield was practically the same, with the exception of group 4, which presented the highest productivity.
Genotypes that presented the lowest values for mature plant height are related to a shorter cycle, because of the shorter interval of time in the vegetative and reproductive stages. Thus, the habit of undetermined growth in soybean genotypes is an extremely important trait, since it aims to reach higher plant height when mature, even in more precocious cycle genotypes (MARQUES; ROCHA; HAMAWAKI, 2008MARQUES, M. C.; ROCHA, J.; HAMAWAKI, O. T. Avaliação do teor de óleo de cultivares de soja da UFU visando a produção de biodiesel. Horizonte Científico, v. 2, n. 1, p. 207-219, 2008.).
Groups 4 and 5 exhibited the most traits dispersion and, consequently, greater genetic variability among genotypes. They can be used in new hybridizations to form segregating populations with the objective of uniting, in one individual, precocity, taller mature plants, as well as larger yield component values. Consequently, the plant would exhibit better total soybean yield. The other genotype groups presented smaller variations since the average soybean yield was similar.
The cluster analysis helps the genetic enhancer to choose a specific group of genotypes according to the desired trait, which enables future use of the same genotypes in commercial cultures or even for composing hybridizations.
Similar results from multivariate analyses were obtained by Vianna et al. (2013)VIANNA, V. F. et al. The multivariate approach and influence of characters in selecting superior soybean genotypes. African Journal of Agricultural Research, v. 8, p. 4162-4169, 2013. while studying the influence of agronomic traits in the selection of genotypes resistant to soybean rust, and by Dallastra et al. (2014)DALLASTRA, A. et al. Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014., while studying soybean progenies selection in segregating populations descending from RR genitors. Furthermore, Barbosa et al. (2013)BARBOSA, R. M., et al. Discrimination of soybean seed lots by multivariate exploratory techniques. Journal of Seed Science, v. 35, n. 3, p. 302-310, 2013., concluded that both multivariate cluster and principal components analyses are efficient tools for describing seed lots.
Interestingly, the results obtained from the Ward hierarchical grouping method produced by the dendrogram were similar to the results obtained by the K-means non-hierarchical method (centroid profile). In addition, the grouping analyses results were similar to those obtained by the principal components analysis, because it classified highly specific genotypes, selected for the aforementioned traits, into the same group. This is proof of the similar results and efficiency of the multivariate exploratory techniques in selecting soybean genotypes with good attributes.
The index developed by Mulamba and Mock (1978)MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978. was used on advanced generations of enhancement to predict the gains enabled by selection. Selection intensity of 19% was applied with the intention of selecting 16 soybean genotypes, i.e., the same quantity selected by the principal components analysis, thus, comparing the results of both selection methods.
Seven RR soybean genotypes were selected by the Mulamba and Mock index (1978)MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978.: 11; 20; 22; 23; 28; 29; 30; 32; 36; 64; 65; 78; 82; 83; and 84. These results were in agreement with those obtained by the multivariate analysis for genotypes: 11. 20; 22; 28; 29; 30; 32; and 36, therefore, there is conformity among these methods, which attests to their efficiency in selecting soybean genotypes. The traits that exhibited the greatest gains after the selection were: number of pods (41.51%), number of branches (19.02%), and grain yield (16.03%) (Table 4). Thus, selection based on some soybean production primary components (number of branches and pods) enable the acquisition of genotypes with higher grain yield by means of indirect selection.
Estimations of selection gain (SG) obtained by the Mulamba and Mock method (1978), in F5 RR soybean genotypes for the traits: NDM - Number of day to maturity (days); PHM - Plant height at maturity (cm), L - Lodging (visual score); AV - Agronomic value (visual score); NB - Number of branches (un); NP - Number of pods (un); WHS - Weight of 100 seeds (g) and GY - Grain yield (kg ha-1)
The Mulamba and Mock (1978)MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978. sum of the rank index is a good method because it contributes to better total gains for the evaluated traits regarding soybean yield, thus, this selection strategy was proven to be efficient in genotype selection. According to Costa et al. (2004)COSTA, M. M. et al. Ganho genético por diferentes critérios de seleção em populações segregantes de soja. Pesquisa Agropecuária Brasileira, v. 39, n. 11, p. 1095-1102, 2004., the index based on the Mulamba and Mock (1978)MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978. sum of ranks was proper and had superior soybean gains in several studied situations. Leite et al. (2016)LEITE, W. S. et al. Estimativas de parâmetros genéticos, correlações e índices de seleção para seis caracteres agronômicos em linhagens F8 de soja. Comunicata Scientiae, v. 7, n. 3, p. 302-310, 2016. and Rosado et al. (2012)ROSADO, L. D. S. et al. Simultaneous selection in progenies of yellow passion fruit using selection indices. Revista Ceres, v. 59, n. 1, p. 95-101, 2012. also confirmed that this index was adequate because it promoted a balanced gain distribution and because it promoted higher total genetic gains by selecting superior genotypes.
Bizari et al. (2017)BIZARI, E. H. et al. Selection indices for agronomic traits in segregating populations of soybean. Revista Ciência Agronômica, v. 48, n. 1, p. 110-117, 2017. evaluated the selection indexes for the agronomic traits in segregating soybean population and concluded again that the Mulanba & Mock sum of ranks index, using the soybean yield and agronomic value as main estimators, provided better gains in their study than the several other applied indexes.
CONCLUSIONS
-
The principal components analysis enabled us to describe and select 16 Roundup Ready soybean genotypes with agronomic superiority;
-
The traits that make up the yield components (number of branches and pods) exert more influence on soybean yield for being positively associated;
-
The K-means and Ward grouping analyses exhibited similar results since they grouped together the specific genotypes for the traits selected by the principal components analysis and put them into the same group;
-
The multivariate analysis selected the same genotypes as the Mulamba and Mock selection index.
-
1
Parte da Dissertação de Mestrado do primeiro autor apresentada ao Programa de Pós-Graduação em Agronomia (Genética e Melhoramento de Plantas) da Universidade Estadual Paulista/UNESP, Campus de Jaboticabal
REFERENCES
- ALCANTARA NETO, F. et al Análise de trilha do rendimento de grãos de soja na microrregião do Alto Médio Gurguéia. Comunicata Scientiae, v. 2, n. 2, p. 107-112, 2011.
- ALMEIDA, R. D.; PELUZIO, J. M.; AFFERRI, F. S. Correlações fenotípicas, genotípicas e ambientais em soja cultivada sob condições várzea irrigada, sul do Tocantins. Bioscience Journal, v. 26, n. 1, p. 95-99, 2010.
- BARBOSA, R. M., et al Discrimination of soybean seed lots by multivariate exploratory techniques. Journal of Seed Science, v. 35, n. 3, p. 302-310, 2013.
- BERTINI, C. H. C. M. et al Análise multivariada e índice de seleção na identificação de genótipos superiores de feijão-caupi. Acta Scientiarum. Agronomy, v. 32, n. 4, p. 613-619, 2010.
- BIZARI, E. H. et al Selection indices for agronomic traits in segregating populations of soybean. Revista Ciência Agronômica, v. 48, n. 1, p. 110-117, 2017.
- CÉLERES. Acompanhamento da adoção de biotecnologia agrícola no Brasil: 2º levantamento grãos safra 2016/17. Dezembro 2016. Available at: <http://www.celeres.com.br/2o-levantamento-de-adocao-da-biotecnologia-agricola-no-brasil-safra-201617/ Accessed on July 07, 2017.
» http://www.celeres.com.br/2o-levantamento-de-adocao-da-biotecnologia-agricola-no-brasil-safra-201617/ - COMPANHIA NACIONAL DE ABASTECIMENTO. Acompanhamento da safra brasileira de grãos: 9º levantamento grãos safra 2016/17. Junho 2017. Available at: <http://www.conab.gov.br/OlalaCMS/uploads/arquivos/17_06_08_09_02_48_boletim_graos_junho_2017.pdf>. Accessed on July 07, 2017.
» http://www.conab.gov.br/OlalaCMS/uploads/arquivos/17_06_08_09_02_48_boletim_graos_junho_2017.pdf - COSTA, M. M. et al Ganho genético por diferentes critérios de seleção em populações segregantes de soja. Pesquisa Agropecuária Brasileira, v. 39, n. 11, p. 1095-1102, 2004.
- CRUZ, C. D. Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum. Agronomy, v. 35, n. 3, p. 271-276, 2013.
- DALLASTRA, A. et al Multivariate approach in the selection of superior soybean progeny which carry the RR gene. Revista Ciência Agronômica, v. 45, n. 3, p. 588-597, 2014.
- FEDERER, W. T. Augmented (or hoonuiaku) designs. Hawaiian Planters Record, v. 55, n. 1, p. 191-208, 1956.
- FERREIRA, J. A. et al Diversidade genética em linhagens avançadas de soja oriundas de cruzamentos biparentais, quádruplos e óctuplos. Revista Ciência Agronômica, v. 46, n. 2, p. 339-351, 2015.
- FERREIRA, M. E.; GRATTAPAGLIA, D. Introdução ao uso de marcadores RAPD e RFLP em análise genética Brasília: Emprapa-Cenargen, 1995. 220 p. (EMBRAPA-CENARGEN. Documentos, 20).
- GONÇALVES, A. et al Estimating combining ability in popcorn lines using multivariate analysis. Chilean Journal of Agricultural Research, v. 74, n. 1, p. 10-15, 2014.
- HAIR, J. F. et al Análise multivariada de dados 6. ed. Porto Alegre: Bookman, 2009. 688 p.
- LEITE, W. S. et al Estimativas de parâmetros genéticos, correlações e índices de seleção para seis caracteres agronômicos em linhagens F8 de soja. Comunicata Scientiae, v. 7, n. 3, p. 302-310, 2016.
- MARCELINO, F. C.; GUIMARÃES, M. F. M.; BARROS, E. G. Detecção e quantificação de alimentos geneticamente modificados: o panorama brasileiro. Revista Ceres, v. 54, n. 313, p. 239-249, 2007.
- MARQUES, M. C.; ROCHA, J.; HAMAWAKI, O. T. Avaliação do teor de óleo de cultivares de soja da UFU visando a produção de biodiesel. Horizonte Científico, v. 2, n. 1, p. 207-219, 2008.
- MATSUO, E. et al Resistance of soybean genotypes to glyphosate. Planta Daninha, v. 27, p. 1063-1073, 2009. Número especial.
- MENEGATTI, A. L. A.; BARROS, A. L. M. D. Análise comparativa dos custos de produção entre soja transgênica e convencional: um estudo de caso para o Estado do Mato Grosso do Sul. Revista de Economia e Sociologia Rural, v. 45, n. 1, p. 163-183, 2007.
- MULAMBA, N. N.; MOCK, J. J. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egyptian Journal of Genetics and Cytology, v. 7, n. 1, p. 40-51, 1978.
- NOGUEIRA, A. P. O. et al Análise de trilha e correlações entre caracteres em soja cultivada em duas épocas de semeadura. Bioscience Journal, v. 28, n. 6, p. 877-888, 2012.
- PELUZIO, J. M. et al Genetic divergence among soybean cultivars in irrigated lowland in the State of Tocantins. Ciência Rural, v. 42, n. 3, p. 395-400, 2012.
- REZENDE, J. C. et al Genetic progress in coffee progenies by different selection criteria. Coffee Science, v. 9, n. 3, p. 347-353, 2014.
- ROSADO, L. D. S. et al Simultaneous selection in progenies of yellow passion fruit using selection indices. Revista Ceres, v. 59, n. 1, p. 95-101, 2012.
- SILVA, F. M. et al Toothpick test: a methodology for the detection of RR soybean plants. Revista Ciência Agronômica, v. 46, n. 2, p. 436-442, 2015.
- SILVA, M. O. et al Discriminação de diferentes classes de solos irrigados com águas salinas, na região de Mossoró (RN), com o uso de análise multivariada. Ambiência, v. 6, n. 2, p. 261-270, 2010.
- STATSOFT. Statistica: data analysis software system, version 10. Tulsa, 2004. Available at: <http://www.statsoft.com>. Accessed on July 25, 2015.
» http://www.statsoft.com - VIANNA, V. F. et al The multivariate approach and influence of characters in selecting superior soybean genotypes. African Journal of Agricultural Research, v. 8, p. 4162-4169, 2013.
- VILLELA, O. et al Genetic divergence of roundup ready (RR) soybean cultivars estimated by phenotypic characteristics and molecular markers. African Journal of Biotechnology, v. 13, n. 26, p. 2613-2625, 2014.
Publication Dates
-
Publication in this collection
Jul-Sep 2018
History
-
Received
07 Dec 2015 -
Accepted
23 Aug 2017