Abstracts
The chloroplast DNA of Phaseolus vulgaris L. vr. Rio Negro was isola ted from chloroplasts obtained by descontiuous sucrose gradient centrifugation. The restriction analysis with the enzymes HindIII, EcoRI and BamHI and their combination, allowed to identified more than 20 fragments of 18 to 0.65kb. The size of Phaseolus vulgaris L. cp DNA was estimated in 140kb with the presence of a repeat sequence of about 22kb.
chloroplast genome; common bean; restriction analysis
O DNA cloroplástico do cultivar Rio Negro (Phaseolus vulgaris L.) foi isolado a partir de cloroplastos obtidos por gradiente descontínuo de sacarose. A análise de restrição com as enzimas HindIII, EcoRI e BamHI e a combinação destas, permitiu a identificação de mais de 20 fragmentos na faixa de 18 a 0.65kb. O tamanho do cp DNA de Phaseolus vulgaris L. foi estimado em 140kb com a existência de sequências repetidas de aproximadamente 22kb.
genoma cloroplástico; feijão; análise de restrição
RESTRICTION ENZYME ANALYSIS OF THE CHLOROPLAST DNA OF Phaseolus vulgaris L. vr. Rio Negro
ANÁLISE DE RESTRIÇÃO DO DNA CLOROPLÁSTICO DE Phaseolus vulgaris vr. Rio Negro
Sergio Echeverrigaray1 1 Professor do Departamento de Ciências Biológicas e Pesquisador do Instituto de Biotecnologia, Universidade de Caxias do Sul, Rua Francisco Getúlio Vargas, 1130, Caixa Postal 1352, 95001-970 - Caxias do Sul, RS. Autor para correspondência. Maria Tereza Vitral Carvalho2 1 Professor do Departamento de Ciências Biológicas e Pesquisador do Instituto de Biotecnologia, Universidade de Caxias do Sul, Rua Francisco Getúlio Vargas, 1130, Caixa Postal 1352, 95001-970 - Caxias do Sul, RS. Autor para correspondência. Eric Derbyshire
- NOTE -
SUMMARY
The chloroplast DNA of Phaseolus vulgaris L. vr. Rio Negro was isola ted from chloroplasts obtained by descontiuous sucrose gradient centrifugation. The restriction analysis with the enzymes HindIII, EcoRI and BamHI and their combination, allowed to identified more than 20 fragments of 18 to 0.65kb. The size of Phaseolus vulgaris L. cp DNA was estimated in 140kb with the presence of a repeat sequence of about 22kb.
Key words: chloroplast genome, common bean, restriction analysis.
RESUMO
O DNA cloroplástico do cultivar Rio Negro (Phaseolus vulgaris L.) foi isolado a partir de cloroplastos obtidos por gradiente descontínuo de sacarose. A análise de restrição com as enzimas HindIII, EcoRI e BamHI e a combinação destas, permitiu a identificação de mais de 20 fragmentos na faixa de 18 a 0.65kb. O tamanho do cp DNA de Phaseolus vulgaris L. foi estimado em 140kb com a existência de seqüências repetidas de aproximadamente 22kb.
Palavras-chave: genoma cloroplástico, feijão, análise de restrição.
Chloroplast DNA restriction fragment pattem analysis has become an invaluable tool in the study of organelle segragation, cytoplasmic mutations, variability of organelle DNA from tissue culture-derived plants, chioroplast DNA recombination, nuclear chloroplast genome interactions and plant taxonomy (CRAWFORD, 1990).
The higher plant chloroplast genome consist of a circular molecule of 110 to 180kb in length, which contains a large inverted repeat sequence of 20-25kb. The only exceptions to this pattem só far reported are pea and broad bean (Vicia faba L.), both members of the legume family (PALMER & THOMPSON, 1981).
Chloroplast DNA has been purified on cesium chloride gradients, by phenol extractions prior to restriction enzyme digestion or by direct in-organelle digestion (KUT & FLICK, 1986). All this procedures are time consuming, require many grams of leaf material or are not adapted for all the species. The present paper describe a method for the rapid extraction of chioroplast DNA using small amounts of plant material and which has been used with success in the isolation and restriction analysis of cpDNA from common bean, maize, soja bean and lettuce. This paper also show the restriction pattems and size of the chloroplast DNA of Phaseolus vulgaris L. var. Rio Negro.
The chloroplast purification was similar to that described by PALMER (1982). Plants grown in the green house were placed in the dark for 24h to reduce the starch content of the leaves. The leaves (40g) were homogenized in a blender with 200ml of extraction buffer (100mM Tris-HCl pH 8.0, 0.35M Sorbitol, 5mM EDTA and 1% p-mercaptoethanol) at 4°C. The homogenate was filtered and centrifuged at 7000xg for 10min at 4°C. The chloroplasts were washed in 60ml of the extraction buffer and resuspended in 7ml of the same buffer. These were layered onto 8ml sucrose gradients (4ml of 40% sucrose under 4ml of 20% sucrose) and centrifuged at 5500xg for 45min at 4°C in a swinging bucket rotor. The chloroplast band from the 20-40% sucrose interphase was removed and washed in 20ml of extraction buffer. The chloroplasts were collected by centrifugation at 6000xg for 10min at 4°C and resuspended in 4ml of the extraction buffer.
For Chloroplast DNA isolation it was added 1ml of Sarkosil (10%) and was incubated at 4° C for 30min and them for 2min at 60°C. For protein extraction and nucleic acids precipitation were used the conventional phenol and ethanol treatments. The RNA were eliminated by RNase digestion.
HindIII, EcoRI and BamHI (Pharmacia) were used to digested 5μg of cpDNA in 50μ1 reactions following the manufacturer specifications. The restricted DNA samples were loaded onto horizontal agarose gels (0.8% agarose in Tris-EDTA-borate buffer) and electrophoresed at 80V for 2h, stained with ethidium bromide and observed under UV light. All gels included marker tracks consisting of the digested fragments of l bacteriophage DNA cleaved with EcoRI and HindIII.
Several systems were tested for the isolation and purification of common bean chloroplast DNA. The miniprep system described by CLARK & HANSON (1983) and the maxiprep system described by KOLODNER & TEWARI (1975) gave nuclear DNA smearing on agarose gels. In general, residual smearing did not permit ctean resolution of the highest molecular weight fragments. The in-organelle DNA digestion system (KUT & FLICK, 1986) gave poor cpDNA yields and low repetibility. The disruption of chloroplasts by Triton-X-100 reduced the overall productivity. The best results were obtained with the new method described in this paper, with a recovery of 25 to 40μg of cpDNA from 40g of common beans leaf material and with a degree of purity that allowed the restriction analysis with all the tested enzymes.
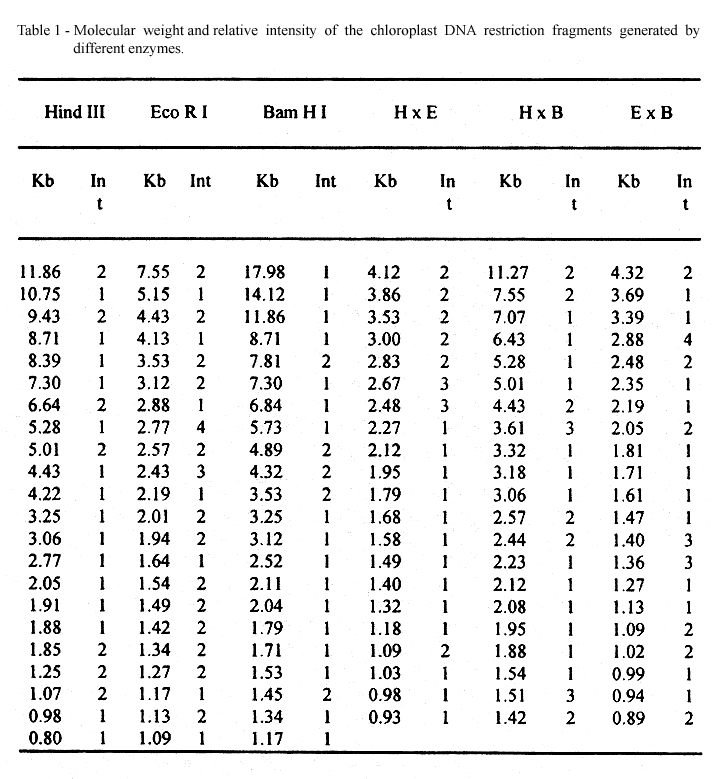
The Phaseolus vulgaris vr. Rio Negro cpDNA was clived with HindIII, EcoRI and BamHI as well as, the combination of these enzymes. The restriction fragments generated by these digestions gave pattems with a high number of bands, in the range of 18 to 0.8kb (Figure 1). As reported by BEDBROOK & KOLODNER, 1979 most of the restriction enzymes have more than 20 cleavege sites in higher plant cpDNA. Comparing with the mobility of the markers (l/HindIII) it was possible to stablished the molecular weigth of each fragment. The visual and densitometric comparison between the intensity of the neightbord bands allowed to estimate the number of fragments that form each band. In Table 1, they can be observed a high number of doble intensity fragments that yield new doble intensity ones when cutted with an other enzyme, thus suggesting the existence of repetitive sequences. The summatory of the molecular weigth of the bands allowed to estimate the size of the Phaseolus vulgaris L. chloroplast DNA in about 140kb with a repeat sequence of about 22kb. The analysis of the chloroplast DNA of other varieties and related species confirm these data. These results are within the range of cpDNA molecular weight for most higher plants (134 to 150kb) and their repeat sequence (20 to 25kb) (BEDBROOK & KOLODNER, 1979).
2 Pesquisador(a) da Seção de Proteínas Vegetais, Centro de Energia Nuclear na Agricultura, Universidade de São Paulo.
Recebido para publicação em 27.02.96. Aprovado em 24.07.96
- BEDBROOK, J.R., KOLODNER. R. The structure of chloroplast DNA. Ann Rev Plant Physiol, v. 30, p. 593-620, 1979.
- CLARK, E.M., HANSON, M.R. Analysis of chloroplast genomes in small amounts of tissue. Plant Molec Biol Rep, v. 1, p. 77-79, 1983.
- CRAWFORD, D.J. Plant molecular systematics: molecular approaches New York: John Wiley & Sons Inc., 1990.
- KOLODNER, R., TEWARI, K.K. The molecular size and conformation of the chloroplast DNA from higher plants. Biochim Biophys Acta, v. 402, p. 372-390, 1975.
- KUT, S.A., FLICK, C.E. A miniprep system for analysis of chloroplast DNA restriction enzyme digest fragments in Nicotina Plant Molec Biol Rep, v. 4, p. 48-55, 1986.
- PALMER, J.E. Physical and gene mapping of chloroplast DNA from Atriplex triangularis and Cucumis sativa Nuc Acids Res, v. 10, p. 1593-1605, 1982.
- PALMER, J.D., THOMPSON, W.F. Rearrengment in the chloroplast genome of mung bean and pea. Proc Natl Acad Sci USA, v. 78, 5533-5537, 1981.
Publication Dates
-
Publication in this collection
24 Sept 2008 -
Date of issue
Dec 1996
History
-
Received
27 Feb 1996 -
Accepted
24 July 1996