ABSTRACT
Bovine digital dermatitis (BDD) is an infectious and contagious disease characterized by ulcerative and proliferative lesions affecting the skin on the bulbs of the heel or the interdigital cleft in dairy cattle, often associated with lameness. Evidences on the etiology of BDD indicate that it is multifactorial, involving environmental factors and multiple bacterial colonization. We isolated and identified microorganisms from BDD biopsy samples obtained from five Holstein Friesian and two Jersey cows by cultivation and molecular identification of bacterial isolates using 16S rRNA gene sequence analysis. We identified six bacterial species: Spirochetes as Treponema pedis and Leptospira broomi/L. fainei, L. licerasiae/L. wolffii; Corynebacterium appendicis, Cupriavidus gilardii and Enterococcus casseliflavus/E. gallinarum. It was quite surprising to have isolated and identified Leptospira species in three out of seven cultures, from different individual cows and two different farms. The species identified belong to the intermediate pathogenic clade, which is a group found to cause human and animal disease. Our findings indicate the need to further investigate the association of Leptospira of intermediate pathogenicity with BDD lesions and whether its presence would have any veterinary and medical significance both in Leptospirosis and with the pathogenesis of BDD lesions, especially in tropical countries
Keywords:
bovine digital dermatitis; molecular identification; 16S rRNA gene analysis; Treponema; Leptospira
RESUMO
Dermatite digital bovina (DDB) é uma doença infecciosa, contagiosa, caracterizada por lesões ulcerativas e proliferativas da região dos talões e/ou do espaço interdigital, frequentemente associada com claudicação. Evidências indicam que a etiologia da DDB é multifatorial, envolvendo fatores ambientais e colonização polimicrobiana. Relata-se aqui o isolamento e a identificação bacteriana em amostras de biópsias em lesões de DDB, obtidas de cinco vacas da raça Holandesa e duas da raça Jersey, por meio de cultivo e identificação molecular de isolados, com base na análise de sequências de genes 16S rRNA. São identificadas seis espécies bacterianas: as espiroquetas Treponema pedis e Leptospira broomi/L. fainei, L. licerasiae/L. wolffii; Corynebacterium appendicis, Cupriavidus gilardii e Enterococcus casseliflavus/E. gallinarum. O isolamento e a identificação de espécies de Leptospira surpreenderam, destacando-se sua presença em três dos sete cultivos obtidos em diferentes vacas, de duas fazendas distintas. As espécies identificadas pertencem ao grupo tipificado como de patogenicidade intermediária, causador de doenças em animais e no homem. Os resultados apresentados indicam a necessidade de maiores investigações sobre a associação entre Leptospira de patogenicidade intermediária e a patogênese das lesões DDB, investigando-se sua presença e significado nas medicinas veterinária e humana, especialmente em países tropicais.
Palavras-chave:
dermatite digital bovina; identificação molecular; análise do gene 16S rRNA; Treponema; Leptospira
INTRODUCTION
Bovine Digital Dermatitis (BDD), or Mortellaro’s disease (Krull et al., 2015KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.), is an infectious and contagious disease characterized by ulcerative and proliferative lesions of the foot skin in dairy cattle and is often associated with lameness. The disease can result in economic losses having a negative impact on reproductive efficiency, productive life span, welfare, and milk production (Walker et al., 1995WALKER, R.L.; READ, D.H.; LORETZ, K.J.; NORDHAUSEN, R.W. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol., v.47, p.343-355, 1995., Fidler et al., 2012FIDLER, A.P.; ALLEY, M.L.; SMITH, G.W. Evaluation of Serpens species bacterin for treatment of digital dermatitis in dairy cattle. Res. Vet. Sci., v.93, p.1258-60, 2012.). BDD has been reported in several European countries, Canada, United States, Mexico, Japan, and in some South American countries like Chile and Brazil (Molina et al., 1999MOLINA, L.R.; CARVALHO, A.U.; FACURY FILHO, E.J. et al. Prevalência e classificação das afecções podais em vacas lactantes na bacia leiteira de Belo Horizonte. Arq. Bras. Med. Vet. Zootec.,v.51, p.149-152, 1999., Nascimento et al., 2015NASCIMENTO, L.V.; MAUERWERK, M.T.; DOS SANTOS, C.L. et al. Treponemes detected in Digital Dermatitis lesions in Brazilian dairy cattle and possible host reservoirs of infection. J. Clin. Microbiol . , v.53, p.1935-1937, 2015.).
The etiology of BDD is not well established, but it is believed to be multifactorial, involving environmental management and physiological and microbiological causes (Klitgaard et al., 2008KLITGAARD, K.; BOYE, M.; CAPION, N.; JENSEN, T.K. Evidence of Multiple Treponema Phylotypes Involved in Bovine Digital Dermatitis as Shown by 16S rRNA Gene Analysis and Fluorescence In Situ Hybridization. J. Clin. Microbiol., v.46, p.3012-3020, 2008.; Zinicola et al., 2015ZINICOLA, M.; LIMA, F.; LIMA, S. et al. Altered Microbiomes in bovine Digital Dermatitis lesions, and the gut as a pathogen reservoir. PLoS ONE, v.10, p.e 0120504, 2015.). Previous studies have demonstrated that multiple bacterial species are associated with lesions, with spirochetes being the most reliably identified organisms (Krull et al., 2015KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.). However, the task of clarifying the etiology of BDD is very difficult because those spirochetes are notoriously difficult to cultivate. Consequently, culture-independent phylogenetic methods, such as comparative 16S rRNA gene sequence analysis of BDD lesions, are a valuable instrument in determining the diversity, phylogeny, prevalence and spatial distribution of unclassified BDD spirochetes (Klitgaard et al., 2008KLITGAARD, K.; BOYE, M.; CAPION, N.; JENSEN, T.K. Evidence of Multiple Treponema Phylotypes Involved in Bovine Digital Dermatitis as Shown by 16S rRNA Gene Analysis and Fluorescence In Situ Hybridization. J. Clin. Microbiol., v.46, p.3012-3020, 2008.). Nevertheless, this method is unable to provide specific information about the physiology of these microorganisms, meaning that it is necessary to make use of culture-dependent approaches.
In this work, we evaluate the presence of bacterial populations, with emphasis on the spirochetes by cultivation, isolation and molecular identification of strains from dairy-cow lesions spontaneously affected by BDD.
MATERIALS AND METHODS
Biopsy samples were obtained from five Holstein Friesian and two Jersey cows with BDD lesions. The sample collection was performed by a veterinarian in two dairy farms around of the city of Belo Horizonte, Minas Gerais state, Brazil. The feet of cows were washed with water and scrubbed with a soft brush to remove soil and other particles attached to the skin surface. Lidocaine 2% (5mL) was injected subcutaneously around the lesion site and skin biopsy specimens, fragments of 6mm, were taken from ulcerative areas of each lesion, using a sterile biopsy punch. The biopsy samples were placed in 2mL of sterile pre-reduced peptone yeast glucose broth (PRAS-PYG) and anaerobically transported to the laboratory. After the biopsies were collected, the seven cows were treated by the veterinarian. Research was approved by the Ethics Committee in Animal Experimentation of Universidade Federal de Minas Gerais (CEUA/UFMG), registration number 121 in 2015.
To verify the presence of microorganisms, the direct examination of biopsy was conducted by observing Gram-stained smears from the broth and, for spiraled microorganisms, observations were made through dark field microscopy (Walker et al., 1995WALKER, R.L.; READ, D.H.; LORETZ, K.J.; NORDHAUSEN, R.W. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol., v.47, p.343-355, 1995.).
Next, samples were mechanically dispersed and homogenized in PRAS-PYG broth, with 200µL of this suspension being used to inoculate 5.0mL of anaerobic oral Treponema isolation broth (OTI) with rifampin 1µg/mL and enrofloxacin 5µg/mL and to inoculate 5.0mL of Leptospira Medium Base Ellinghausen and McCullough modified by Johnson and Harris (EMJH) (Difco, BD Biosciences), supplemented with bovine albumin (Johnson and Faine, 1984JOHNSON, R.C.; FAINE, S. Leptospira, In: KRIEG, N.R.; HOLT, J.G. (Eds.). Bergey's manual of systematic bacteriology. Baltimore:Williams & Wilkins, 1984. p.62-67.) plus 5µg/mL enrofloxacin. To cultivate Leptospira, incubation was at 28ºC and 37ºC for up to 4 weeks in microaerophilic and anaerobic conditions. When initial growth was perceived, other replicas were made in EMJH medium.
DNA was isolated from cultivated bacteria. The entire cultures of each sample (3mL) were centrifuged (8000 x g, 10min) and genomic DNA was obtained using the Wizard Genomic DNA Purification Kit (Promega) according to recommendations of the manufacturer.
Species-level identification was performed by 16S rRNA gene sequencing and sequencing analysis. PCR reactions were performed in 50µL total volume containing 30 ρmols of each 27F (5' AGA GTT TGA TCC TGG CTC AG 3') and 1492R (5' GGT TAC CTT GTT ACG ACT T 3') primers, described by Lane (1991LANE, D.J. 16S/23S rRNA sequencing. In: STACKEBRANDT, E.; GOODFELLOW, M. (Eds.). Nucleic acid techniques in bacterial systematics. Chichester: John Wiley and Sons, 1991. p.115-175.), 10ng of DNA template and PCR Master Mix (Promega Corporation, Madison, USA) with 200µM dNTPs, 1.5mM MgCl2 and 1.5 units Taq DNA polymerase. Amplification occurred over 35 cycles (95°C for 30 s, 55°C for 1 min and 72°C for 1 min) after an initial denaturation (95°C for 2 min) and completed by a final extension (72°C for 5 min). Negative and positive controls were included. The amplicons were purified using the Wizard® SV Gel and PCR Clean-up System kit (Promega Corporation, Madison, USA) and sequenced using the DYEnamic ET Dye Terminator Cycle Sequencing Kit (GE Healthcare, Piscataway, USA) for the system automatic sequencing of MegaBACE 1000 (GE Healthcare, Piscataway, USA).
Forward and reverse sequence reads were assembled using the Phred-Phrap-Consed package with default parameters. The consensus sequence obtained was compared with those deposited in the GenBank database using the BLASTn algorithm v.2.215, available on the website of the National Center for Biotechnology Information (http://www.ncbi/nlm/nih.gov/blast), to find a match to nearly identical sequences (>97% identity) for the 16S rRNA.
The phylogenetic analyses were performed using MEGA 6.06 software with default parameters (Tamura K. et. al 2013TAMURA, K.; STECHER, G.; PETERSON, D. et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. , v.30, p.2725-2729, 2013.). Dataset sequences of 16S rRNA of type strains belonging to Corynebacterium, Cupriavidus, Enterococcus, Leptospira and Treponema genera were obtained from GenBank (supplementary material) and used to perform a multiple sequence alignment using ClustalW software. Next, the best mutational model was calculated for each genus, which was used as a parameter for phylogenetic inferences (evolutionary model of nucleic acids) by the Maximum likelihood method (Hall, 2013HALL, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol., v.30, p.1229-1235, 2013.).
RESULTS
Preparations of all biopsies were examined in dark-field microscopy for the presence of spirochetes, and smears were also observed through light microscopy after Gram staining before and after cultivating. Spirochetes were observed in four biopsy suspensions (A8T, 6MC, 7MC, and 8MC). In addition, short rods (4MC, 9MC) and cocci (5MC) were detected (Tab. 1)
Molecular identification of bacterial isolates using 16S rRNA gene sequencing and sequences analysis
All seven biopsy samples became positive in the cultures. The 16S rRNA gene PCR amplified products from each of the seven bacterial isolates were sequenced. The partial nucleotide sequences obtained (average of 760 base pairs) were analyzed by BLASTN searches in order to compare our consensus sequences against specific 16S ribosomal RNA (Bacteria and Archaea) database at the NCBI website. The number of nucleotides sequenced, the highest similarities found for each sample and the most probable species are indicated in (Tab. 1).
Pure spirochete cultures were obtained from four cows. Comparison of the bacterial 16S rRNA gene sequences from samples A8T, 7MC and 8MC by BLASTN searches showed 99% identity with members of the Leptospira genus. The sequences from samples A8T and 8MC had best hits with L. broomi and L. fainei serovar Hurstbridge. The sequence from sample 7MC had best hits with L. licerasiae serovar Varillal and L. wolffii serovar Khorat (Tab. 1).
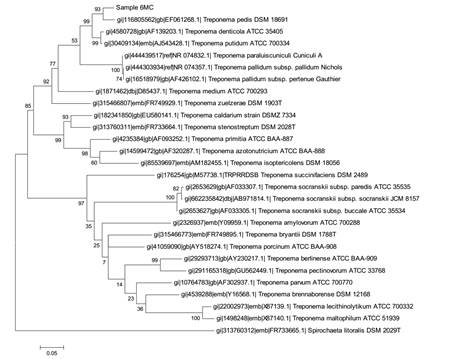
The Figure 1 shows the molecular phylogenetic analysis by maximum likelihood of the Leptospira sequences obtained from pure spirochete cultures. The fourth spirochete positive sample was 6MC and the sequence obtained showed the best hit with Treponema pedis (98%), as confirmed by the molecular phylogenetic analysis shown in Figure 2.
Molecular Phylogenetic analysis by maximum likelihood method of three pure spirochete cultures (samples 7MC, A8T, 8MC) for identification as Lepstospira. The sequences from samples A8T and 8MC had best hits with L. broomi and L. fainei serovar Hurstbridge. The sequence from sample 7MC had best hits with L. licerasiae serovar Varillal and L. wolffii serovar Khorat.
Molecular Phylogenetic analysis by maximum likelihood method for molecular identification of a pure culture sample 6MC as Treponema pedis.
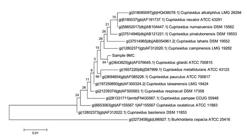
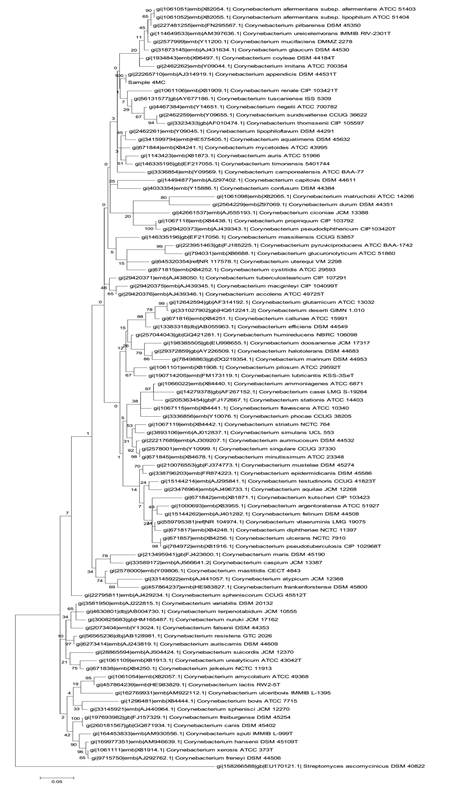
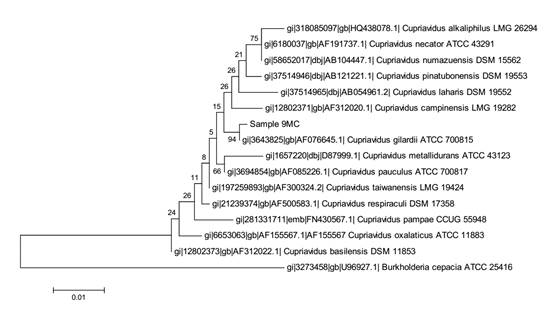
From sample 4MC, the bacterial 16S rRNA gene sequences from pure culture showed the highest similarity with Corynebacterium appendicis (99.5%) and the corresponding phylogenetic tree is presented in Figure 3. From sample 5MC, the sequences with the best hits indicated the genus Enterococcus, and the species Enterococcus casseliflavus and E. gallinarum presenting the highest similarity (99%), as shown in the phylogenetic tree in Figure 4. Also, the sequences obtained from sample 9MC showed the best hits with Cupriavidus gilardii (99%) and the corresponding phylogenetic tree is presented in Figure 5.
Molecular Phylogenetic analysis by Maximum Likelihood method for molecular identification of a pure culture (sample 4MC) as Corynebacterium appendicis .
Molecular Phylogenetic analysis by Maximum Likelihood method for identification of the pure culture corresponding to sample 5MC as Enterococcus casseliflavus and E. gallinarum.
Molecular Phylogenetic analysis by Maximum Likelihood method for identification of the pure culture corresponding to sample 9MC as Cupriavidus gilardii.
DISCUSSION
According to Krull et al. (2015KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.), sequencing of DNA from 48 biopsies confirmed the presence of complex communities composed of one hundred bacterial species present in the lesions. Treponemes and Corynebacteriaceae were among the top 12 bacterial species for each lesion stage. In the present work, both Treponemes and Corynebacteriaceae were isolated and identified from cultures derived from samples 6MC and 4MC (Table 1), respectively, as T. pedis and C. appendicis.Evans et al., 2009EVANS, N.J.; BROWN, J.M.; DEMIRKAN, I. et al. Treponema pedis sp. nov., a spirochaete isolated from bovine digital dermatitis lesions. Int. J. Syst. Evol. Microbiol., v.59, p.987-991, 2009. , describes Treponema pedis sp.nov. a spirochaete representing a novel species of the genus Treponema isolated from bovine digital dermatitis lesions. The novel isolates form a coherent Treponema sp. taxonomic group. In the USA, 99.0% of the samples isolated from BDD lesions were identified as T. pedis sp. nov. strains by 16S rRNA gene sequence similarity, (Walker et al.,1995WALKER, R.L.; READ, D.H.; LORETZ, K.J.; NORDHAUSEN, R.W. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol., v.47, p.343-355, 1995.). This possibly indicates worldwide (UK, USA, Germany ) presence of T. pedis sp.nov. suggesting that this microorganism might have a significant etiological role in BDD. On the other hand, BDD appears to have a complicated etiology involving co-infection by several different treponemes (Choin et al., 1997CHOI, B.K.; NATTERMANN, H.; GRUND, S. et al. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol., v.47, p.175-181, 1997.). The presence of a diverse range of treponemes in polytreponemally infected tissue has been identified by cloning and sequencing of 16S rRNA genes, but their highly fastidious nature has, for the most part, precluded any further characterization of these organisms (Evans et al., 2009EVANS, N.J.; BROWN, J.M.; DEMIRKAN, I. et al. Treponema pedis sp. nov., a spirochaete isolated from bovine digital dermatitis lesions. Int. J. Syst. Evol. Microbiol., v.59, p.987-991, 2009. ).
C. appendicis, a species proposed by Yassin et al. (2002YASSIN, A.F.; STEINER, U.; LUDWIG, W. Corynebacterium appendicis sp.nov. Int. J. Syst. Evol. Microbiol. , v.52, p.1165-1169, 2002.), is a facultative anaerobe that was previously isolated from abdominal swabs of a human patient with appendicitis accompanied with abscess formation. The culture derived from sample 5MC had the best hits with Enterococcus casseliflavus and Enterococcus gallinarum. The Enterococci are Firmicutes, which form part of the intestinal microbiota of most mammals and birds, and have emerged as important nosocomial and community-acquired pathogens (Jung et al., 2007JUNG, W.K.; LIM, J.Y.; KWON, N.H. et al. Vancomycin-resistant enterococci from animal sources in Korea. Int. J. Food. Microbiol., v.113, p.102-107, 2007.). The isolation and identification of Treponeme, Corynecbacterium and Firmicutes is in agreement with previous works that employed molecular identification of microorganisms from BDD lesions (Zinicola et al., 2015ZINICOLA, M.; LIMA, F.; LIMA, S. et al. Altered Microbiomes in bovine Digital Dermatitis lesions, and the gut as a pathogen reservoir. PLoS ONE, v.10, p.e 0120504, 2015.).
The culture derived from sample 9MC (Tab.1) had best hits with Cupriavidus gilardii, a Gram-negative, motile rod that occurs in the environment, soil and human clinical specimens (Vandame and Coeny, 2004). C. gilardii has been isolated from human clinical samples that include respiratory tract infection (cystic fibrosis patients, Lipuma, 2010LIPUMA, J.J. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol.Rev., v.23, p.299-323, 2010. ).
It was quite surprising to have isolated and identified Leptospira species derived from biopsies of BDD lesions. As previously stated, the sequences from samples A8T and 8MC had best hits with L. broomi and L. fainei and that from sample 7MC had best hits with L. licerasiae and L. wolffii (Tab.1). Therefore, in three out of seven cultures, isolated from different individual cows from different farms, we positively identified Leptospira that could belong to four distinct species: L. broomi, L. fainei, L. licerasiae or L. wolffii. We could not find in the literature any previous description of the association of this genus with BDD lesions.
Leptospirosis is a worldwide zoonotic disease caused by infections with pathogenic spirochetes of the genus Leptospira which affects wild and domestic animals and humans. Infected animals excrete the organism in their urine and infect other animals by direct exposure to the contaminated urine or through indirect contact via contaminated water or soil (Zuerner, 2010ZUERNER, R.L. Genus I: Leptospira. Family IV: Leptospiraceae. In: KRIEG, N.R.; STALEY, J.T.; BROWN, D.R.; et al. (Eds.). Bergey's manual of systematic bacteriology , 2.ed. New York: Springer, 2010. v.4, p.546-556.). Comparisons of 16S rRNA sequences and DNA-DNA reassociation studies distinguished the presence of three clades in the genus Leptospira, the first is of pathogenic Leptospira, the second of intermediate pathogenic species and the third is of nonpathogenic species (Levett, 2015LEVETT, P.N. Systematics of leptospiraceae. Curr. Top. Microbiol. Immunol., v.387, p.11-20, 2015.). There are five species with intermediate pathogenicity: L. fainei, L. inadai, L. broomi, L. licerasiae and L. wolffii (Levett, 2015LEVETT, P.N. Systematics of leptospiraceae. Curr. Top. Microbiol. Immunol., v.387, p.11-20, 2015.). It is interesting to mention that all Leptospira isolated in the present work (L. broomii, L. fainei, L. licerasiae or L. wolffii) belong to the intermediate pathogenic clade (Zuerner, 2010ZUERNER, R.L. Genus I: Leptospira. Family IV: Leptospiraceae. In: KRIEG, N.R.; STALEY, J.T.; BROWN, D.R.; et al. (Eds.). Bergey's manual of systematic bacteriology , 2.ed. New York: Springer, 2010. v.4, p.546-556.). These intermediate pathogenic species are found to cause human and animal disease. For instance, L. broomii was isolated from the blood, cerebrospinal fluid and urine of human patients with leptospirosis in Denmark and France (Levett et al., 2006LEVETT, P.N.; MOREY, R.E.; GALLOWAY, R.L.; STEIGERWALT, A.G. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int. J. Syst. Evol. Microbiol. , v.56, p.671-673, 2006.). L. fainei was previously isolated from cultures of uteri and kidneys of sows from two geographically distant pig herds in Australia (Perolat et al., 1998PEROLAT, P.; CHAPPEL, R.J.; ADLER, B. et al. Leptospira fainei sp. nov., isolated from pigs in Australia. Int. J. Syst. Bacteriol. , v.48, p.851-858, 1998. ). L. fainei was successively cultured from two human patients with Weil’s disease and antibody titers have been found in sera from cattle in Australia, France and Indonesia (Chappel et al., 1996CHAPPEL, R.J.; BILLINGHURST, M.L.; ADLER, B. et al. Isolation from pigs of a Pathogenic leptospire of a new Genospecies and serogrup (serovar hurstbridge) and a serological study of its distribution and possible veterinary significance. In: MEETING OF THE INTERNATIONAL LEPTOSPIROSIS SOCIETY, 1., 1996, Nantes. Proceedings... Nantes, France: [s.l.], 1996.; Petersen et al., 2001PETERSEN, A.M.; BOYE, K.; BLOM, J. et al. First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J. Med. Microbiol., v.50, p.96-100, 2001.).
L. wolffii was identified as a potential pathogenic species, detected in infected humans, in sheep sera and dogs urine and can be potentially pathogenic to human and probably animal hosts (Zakeri et al., 2010ZAKERI, S.; KHORAMI, N.; GANJI, Z.F. et al. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect. Genet. Evol., v.10, p.273-277, 2010.); and L. licerasiae causes mild disease in humans and has been isolated from peridomestic and wild rodents and marsupials in Peruvian Amazon (Matthias et al., 2008MATTHIAS, M.A.; RICALDI, J.N.; CESPEDES, M. et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLOS Negl. Trop. Dis., v.2, p.e213, 2008.; Ricaldi et al., 2012RICALDI, J.N.; FOUTS, D.E.; SELENGUT, J.D. et al. Whole genome analysis of Leptospira licerasie provides insight into letpospiral evolution and pathogenicity. PLOS Negl. Trop. Dis. , v.6, p.e1853, 2012.) and from environmental water (Saito et al., 2013SAITO, M.; VILLANUEVA, S.Y.A.M.; CHAKRABORTY, A. et al. Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl. Environ. Microbiol., v.79, p.601-609, 2013. ).
Krull et al. (2015KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.) showed that early BDD lesions had minimal populations of Treponema spp. that grow significantly in more advanced and active lesion stages. In contrast, early lesions were composed of much more complex bacterial communities suggesting that it would have developed as a result of the concurrent presence of many bacterial species at the same time. The authors admitted that these findings fit the hypothesis that the treponemes represent an opportunistic organism that colonizes pre-existing lesions of the foot of dairy cattle. As the Treponeme species fail to have significant genetic capabilities to break down the skin barrier, they likely colonize the hospitable niche associated with skin lesions induced by other bacterial organisms or physical trauma to the foot (Krull et al., 2015KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.). In contrast to Treponema species that fail to have significant genetic capabilities to break down the skin barrier (Krull et al., 2015KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.), L. interrogans, the most common genotype in pathogenic Leptospira species, encodes a collagenase (colA gene), which was recently related to invasiveness and transmission of the spirochetes in cell monolayers and animals, being characterized as a novel and crucial virulence factor (Kassegne et al., 2014KASSEGNE, K.; HU, W.; OJCIUS, D.M. et al. Identification of Collagenase as a Critical Virulence Factor for Invasiveness and Transmission of Pathogenic Leptospira Species. J. Infect. Dis., v.209, p.1105-1115, 2014.). Ricaldi et al. (2012RICALDI, J.N.; FOUTS, D.E.; SELENGUT, J.D. et al. Whole genome analysis of Leptospira licerasie provides insight into letpospiral evolution and pathogenicity. PLOS Negl. Trop. Dis. , v.6, p.e1853, 2012.) sequenced genomes of two strains of L. licerasiae and these had the highest average protein identity with L. interrogans. These observations suggest that L. licerasiae is more closely related to the pathogenic branch of infectious Leptospira than to the saprophyte L. biflexa.
Our results are in general agreement with evidences that point to the polymicrobial nature of the BDD. The unexpected finding of the isolation and identification of Leptospira of intermediate pathogenicity from deep BDD lesion biopsies indicates the need for further investigations in order to clarify whether, besides the treponeme, other spirochetes, as Leptospira, could be associated with BDD and even the probability of its participation in the pathogenesis of disease. Another interesting hypothesis would be that BDD skin lesions would allow the access of Leptospira of intermediate pathogenicity as a route to cause Leptospirosis in cattle, especially in tropical countries, clarifying whether the results of the present work could have any veterinary and medical significance.
CONCLUSION
Multiple bacterial species are associated with BDD lesions. Here we report the isolation and identification of Treponema pedis, Corynebacterium appendicis, Enterococcus casseliflavus/ gallinarum, Cupriavidus gillardi and Leptospira that could belong to four distinct species: L. broomi, L. fainei, L. licerasiae or L. wolffii. Finding Leptospira of intermediate pathogenicity from deep BDD lesion biopsies indicates the need for further investigations in order to clarify whether, besides the treponeme, other spirochetes, as Leptospira, could be associated with BDD and even the probability of its participation in the pathogenesis of disease.
ACKNOWLEDGEMENTS
Our thanks to Brazilian financing scientific programs or institutions: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de nível Superior (CAPES) and Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG), Antônio Ultimo de Carvalho, Elias Facury Filho, Jacques Robert Nicoli, Luciana Márcia de Oliveira, Luiz de Macedo Farias, Paula Prazeres Magalhães and Simone Gonçalves dos Santos, for theorical and technical assistance.
REFERENCES
- CHAPPEL, R.J.; BILLINGHURST, M.L.; ADLER, B. et al. Isolation from pigs of a Pathogenic leptospire of a new Genospecies and serogrup (serovar hurstbridge) and a serological study of its distribution and possible veterinary significance. In: MEETING OF THE INTERNATIONAL LEPTOSPIROSIS SOCIETY, 1., 1996, Nantes. Proceedings... Nantes, France: [s.l.], 1996.
- CHOI, B.K.; NATTERMANN, H.; GRUND, S. et al. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol., v.47, p.175-181, 1997.
- EVANS, N.J.; BROWN, J.M.; DEMIRKAN, I. et al. Treponema pedis sp. nov., a spirochaete isolated from bovine digital dermatitis lesions. Int. J. Syst. Evol. Microbiol., v.59, p.987-991, 2009.
- FIDLER, A.P.; ALLEY, M.L.; SMITH, G.W. Evaluation of Serpens species bacterin for treatment of digital dermatitis in dairy cattle. Res. Vet. Sci., v.93, p.1258-60, 2012.
- HALL, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol., v.30, p.1229-1235, 2013.
- JOHNSON, R.C.; FAINE, S. Leptospira, In: KRIEG, N.R.; HOLT, J.G. (Eds.). Bergey's manual of systematic bacteriology. Baltimore:Williams & Wilkins, 1984. p.62-67.
- JUNG, W.K.; LIM, J.Y.; KWON, N.H. et al. Vancomycin-resistant enterococci from animal sources in Korea. Int. J. Food. Microbiol., v.113, p.102-107, 2007.
- KASSEGNE, K.; HU, W.; OJCIUS, D.M. et al. Identification of Collagenase as a Critical Virulence Factor for Invasiveness and Transmission of Pathogenic Leptospira Species. J. Infect. Dis., v.209, p.1105-1115, 2014.
- KLITGAARD, K.; BOYE, M.; CAPION, N.; JENSEN, T.K. Evidence of Multiple Treponema Phylotypes Involved in Bovine Digital Dermatitis as Shown by 16S rRNA Gene Analysis and Fluorescence In Situ Hybridization. J. Clin. Microbiol., v.46, p.3012-3020, 2008.
- KRULL, A.C.; SHEARER, J.K.; GORDEN, P.J. et al. Bacterial causes of Digital Dermatitis (DD) in dairy cattle. Anim. Ind. Rep., AS 661, ASL R2972, 2015.
- LANE, D.J. 16S/23S rRNA sequencing. In: STACKEBRANDT, E.; GOODFELLOW, M. (Eds.). Nucleic acid techniques in bacterial systematics. Chichester: John Wiley and Sons, 1991. p.115-175.
- LEVETT, P.N. Systematics of leptospiraceae. Curr. Top. Microbiol. Immunol., v.387, p.11-20, 2015.
- LEVETT, P.N.; MOREY, R.E.; GALLOWAY, R.L.; STEIGERWALT, A.G. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int. J. Syst. Evol. Microbiol. , v.56, p.671-673, 2006.
- LIPUMA, J.J. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol.Rev., v.23, p.299-323, 2010.
- MATTHIAS, M.A.; RICALDI, J.N.; CESPEDES, M. et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLOS Negl. Trop. Dis., v.2, p.e213, 2008.
- MOLINA, L.R.; CARVALHO, A.U.; FACURY FILHO, E.J. et al. Prevalência e classificação das afecções podais em vacas lactantes na bacia leiteira de Belo Horizonte. Arq. Bras. Med. Vet. Zootec.,v.51, p.149-152, 1999.
- NASCIMENTO, L.V.; MAUERWERK, M.T.; DOS SANTOS, C.L. et al. Treponemes detected in Digital Dermatitis lesions in Brazilian dairy cattle and possible host reservoirs of infection. J. Clin. Microbiol . , v.53, p.1935-1937, 2015.
- PEROLAT, P.; CHAPPEL, R.J.; ADLER, B. et al. Leptospira fainei sp. nov., isolated from pigs in Australia. Int. J. Syst. Bacteriol. , v.48, p.851-858, 1998.
- PETERSEN, A.M.; BOYE, K.; BLOM, J. et al. First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J. Med. Microbiol., v.50, p.96-100, 2001.
- RICALDI, J.N.; FOUTS, D.E.; SELENGUT, J.D. et al. Whole genome analysis of Leptospira licerasie provides insight into letpospiral evolution and pathogenicity. PLOS Negl. Trop. Dis. , v.6, p.e1853, 2012.
- SAITO, M.; VILLANUEVA, S.Y.A.M.; CHAKRABORTY, A. et al. Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl. Environ. Microbiol., v.79, p.601-609, 2013.
- TAMURA, K.; STECHER, G.; PETERSON, D. et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. , v.30, p.2725-2729, 2013.
- VANDAMME, P.; COENYE, T. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. , v.54, p.2285-2289, 2004.
- WALKER, R.L.; READ, D.H.; LORETZ, K.J.; NORDHAUSEN, R.W. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol., v.47, p.343-355, 1995.
- YASSIN, A.F.; STEINER, U.; LUDWIG, W. Corynebacterium appendicis sp.nov. Int. J. Syst. Evol. Microbiol. , v.52, p.1165-1169, 2002.
- ZAKERI, S.; KHORAMI, N.; GANJI, Z.F. et al. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect. Genet. Evol., v.10, p.273-277, 2010.
- ZINICOLA, M.; LIMA, F.; LIMA, S. et al. Altered Microbiomes in bovine Digital Dermatitis lesions, and the gut as a pathogen reservoir. PLoS ONE, v.10, p.e 0120504, 2015.
- ZUERNER, R.L. Genus I: Leptospira. Family IV: Leptospiraceae. In: KRIEG, N.R.; STALEY, J.T.; BROWN, D.R.; et al. (Eds.). Bergey's manual of systematic bacteriology , 2.ed. New York: Springer, 2010. v.4, p.546-556.
Publication Dates
-
Publication in this collection
June 2017
History
-
Received
11 Oct 2016 -
Accepted
24 Nov 2016